Abstract
The microbiome–gut–brain axis field is multidisciplinary, benefiting from the expertise of microbiology, ecology, psychiatry, computational biology, and epidemiology among other disciplines. As the field matures and moves beyond a basic demonstration of its relevance, it is critical that study design and analyses are robust and foster reproducibility. In this companion piece to Bugs as features (part 1), we present techniques from adjacent and disparate fields to enrich and inform the analysis of microbiome–gut–brain axis data. Emerging techniques built specifically for the microbiome–gut–brain axis are also demonstrated. All of these methods are contextualized to inform several common challenges: how do we establish causality; how can we integrate data from multiple ’omics techniques; how might we account for the dynamicism of host–microbiome interactions? This perspective is offered to experienced and emerging microbiome scientists alike to assist with these questions and others at the study conception, design, analysis, and interpretation stages of research.
Similar content being viewed by others
Main
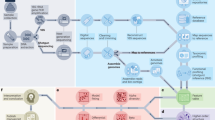
The microbiome–gut–brain axis is informed by biological and epistemological knowledge from many disciplines, spanning microbiology, ecology, psychiatry, and others. Similarly, in its analysis, it is strengthened by methods from across the scientific landscape, as well as some truly interdisciplinary approaches developed specifically for the microbiome–gut–brain axis field (Fig. 1).
a, Constructing a DAG facilitates statistical techniques such as causal inference, and experimental procedures such as FMT can be used to interrogate causality and directionality. b, Techniques such as multivariate modeling and ordination can be used to analyze and interpret large’omics datasets. c, Higher-order patterns within microbiome data such as interaction networks, functional modules, ecological guilds, and amalgamations, called mesoscale features, are used to ask and answer ecologically relevant questions. d, Microbiome time series can be analyzed using mixed-effect models. Special cases of time-series microbiome data, such as microbial volatility, as well as circadian rhythms can be used to ask and answer targeted questions.
In part 1, we introduced core concepts and foundations of compositional data analysis of the microbiome–gut–brain axis:1, ranging from study design and pre-registration of analysis, to selecting the most suitable diversity metrics, and the options for functional inference. In part 2, we provide a perspective on how to leverage techniques from other disciplines, and provide future directions for the microbiome–gut–brain axis field. We hope that this mapping of the broader landscape will provide useful navigation from which the reader may explore original sources as per their needs and interests.
One aim of this piece is to provide context for the methods borrowed, adapted, and developed from both adjacent and far-flung fields and to aid the reader in appraising their respective strengths and weaknesses for microbiome analysis.
As a guiding principle, we believe that the microbiome–gut–brain axis field has an imperative to become a more reproducible science and to operate from a place of deeper statistical and biological understanding. The techniques described in the following have been carefully examined and selected to ensure that they are fit to drive the field toward this goal.
Causality, uncertainty and the microbiome
There has been a growing call for experiments that can establish causality in the microbiome–gut–brain axis field2,3. Causality is a philosophically and statistically contentious term. Granger causality can be thought of as a pragmatic approach to estimating causality between occurrences A and B. In a nutshell, if knowledge of the occurrence A helps predict the occurrence of B, A is said to ‘Granger-cause’ B. However, in the case of complex systems such as the microbiome, where nonlinear dynamics are ubiquitous, Granger causality may not be appropriate4. Historically compelling sets of criteria to establish causality between a microorganism and a disease exist, including Koch’s postulates5 and the Bradford Hill criteria6. Since these are less applicable to the ecosystem approach required for the microbiome–gut–brain axis, we will not elaborate on these criteria (but see Box 1, which leverages experimental design to interrogate causality). Rather, we provide an overview of causality concepts from epidemiology and econometrics that have been applied to the microbiome–gut–brain axis, as well as some important pitfalls.
Causal inference analysis
Causal inference is commonly the underlying motivation for microbiome–gut–brain axis studies, even when it is not being explicitly tested. As outlined by ref. 7, being explicit about the causal motivations of (even) an observational analysis “reduces ambiguity in the scientific question, errors in the data analysis, and excesses in the interpretation of the results”. Rather than avoiding causal language because, as the oft-repeated cautionary tale goes, correlation does not mean causation, Hernán7 suggests that we instead ask clearer causal questions and improve use of causal inference methods such as adjusting for confounding. There are many occasions where randomized controlled trials that would provide stronger evidence of causality are not feasible, biologically plausible, or indeed ethical. The complexity of the gastrointestinal and microbial environments is certainly difficult to replicate completely as interventions in clinical and even preclinical trials.
A directed acyclic graph (DAG8), or causal diagram, is a useful first step in making explicit the causal hypotheses and underlying assumptions about variables in a study. Creating a DAG serves as a prompt to consider, discuss with colleagues, and design analyses. It is best done at the conception phase of a study so that it may inform aspects of study design, from the timing of data collection to the list of potentially confounding variables about which to collect data. We stress here that while DAGs are a helpful tool to ask causal questions, they do not necessarily allow the user to quantify causality from cross-sectional data. Specialized mechanistic follow-up studies remain the gold standard in this regard. In brief, hypothesized relationships between variables are represented by arrows between them, pointing from cause to effect. By convention, causal diagrams point left to right, with exposure variables on the left and outcome variable on the right. Then add any variables that causally impact the main exposure of interest, or the outcome, using arrows between variables to depict the direction of causality. DAGs must be acyclic; that is, variables must not contain feedback loops; relationships between variables must be depicted as unidirectional. This differs from infographics of gut–brain interactions, which are frequently bidirectional as per biological reality. An example DAG can be found in Fig. 2a, and we expand on DAG creation in Box 2. This DAG was created using the dagitty R library and reflects variables relevant to the previously published schizophrenia dataset9 used in the accompanying Rmarkdown script.
a, A DAG describes a hypothetical causal pathway, where an arrow from A to B (A → B) suggests a causal relationship between A and B. Ideally, the framework of constructs and variables is considered before (and thereby informing) data collection. Some examples of relevant unobserved variables are shown here in gray. Under the assumptions of this DAG and with only three available covariates (sex, smoking status, and body mass index), sex and smoking status comprise the minimal adjustment set required to estimate the total effect of gut microbiota on schizophrenia. b, Table illustrating the relationship between models where predictors and responses can be either univariate or multivariate. Predictor variables are on the left of each diagram, whereas response variables are on the right. Univariate predictor variable to multivariate response variables was included for the sake of completion. Practically, a univariate predictor variable to multivariate response variables can be approached in the same fashion as multivariate predictor variable to a univariate response variable. c, Two DAGs illustrating the scenarios from the section on mediation analysis. Diet affects the microbiome, and diet affects the brain. Mediation analysis helps us ask and answer whether measured associations between the microbiome and the brain are spurious (scenario I) or whether the microbiome affects the brain through diet (scenario II). BMI, body mass index.
Mediation analysis
Mediation analysis is used to investigate whether a variable transmits its effect on the outcome through another mediator variable10. For example, an effect of diet on host behavior is well documented, as are effects of diet on the microbiome11. Similarly, the gut microbiome is also known to affect host behavior12. If we were to test whether diet could affect host behavior via its effects on the microbiome, that would require a mediation analysis. Where mediation explains a relationship, there are two main possibilities:
-
Partial mediation refers to the scenario where there is both a direct effect and an indirect (mediation) effect; for example, if diet were to both directly affect behavior and indirectly affect behavior by modulating the microbiome—which in turn affects behavior.
-
Complete mediation refers to the scenario where—using the preceding example again—diet affects only the microbiome, which in turn affects behavior, but diet on its own does not directly affect behavior.
One recent example of how mediation analysis can be used in the microbiome–gut–brain axis field can be found in the context of autism, diet, and the microbiome13. The authors convincingly showed that alterations in the microbiomes of autistic children can be explained by a restricted diet, a common trait in autistic children. They concluded that since diet can explain the altered microbiome, that altered microbiome does not play a causal role in the occurrence of autism. In a letter to Yap et al.13, Morton et al.14 argued that their model implicitly assumed the absence of a relationship between diet and the microbiome (for example, independence), which is known to be untrue. Morton et al.14 argued that a more appropriate model would be one where diet affects (1) host phenotype directly and (2) the microbiome, which in turn affects phenotype. Essentially, they argue that the microbiome acts as a partial mediator in this autism example. See Fig. 2c for two miniature DAGs illustrating these two scenarios.
Several excellent tools exist to perform mediation analysis. The mediation package in R takes standard generalized linear model fits as input15. Also see the primer on how to perform a mediation analysis in R in the Supplementary Information.
We note that mediation is accompanied by inherently longitudinal assumptions. One presumes that due to the occurrence of some exposure at time 1, a mediating variable is affected at time 2, and the outcome shift as a result is observed at some point in the future (time 3). The use of mediation analysis in cross-sectional observational data, although common, is not considered best practice16. The reason for this is that it presumes that the causal chain being tested is correct and precludes an examination of a potential alternative temporal order of the variables. This is particularly relevant for variables that are dynamic, such as diet, the microbiome, and mental states. So mediation analysis in cross-sectional observational studies is correlational and needs to be validated in targeted follow-up studies. Some alternative options with fewer data requirements have been trialed through data simulation,17 demonstrating that sequential mediation—when data for the exposure, mediator, and outcome are collected only once each, but at least longitudinally and in a meaningful temporal sequence—can provide adequate sensitivity to identifying the presence of mediation. The gold standard is the resource-intensive multilevel longitudinal mediation, where variables that represent enduring exposures (such as diet) are collected repeatedly, and path coefficients between variables are allowed to vary across individuals. This may also be ideal for contexts in which a large degree of inter-individual variability might be expected (such as in host–microbiome studies).
Notably, estimating an indirect effect through mediation analysis requires substantially more power than estimating direct effects in traditional analysis. For example, one popular method to estimate the effect size of a mediation analysis is to multiply the two coefficients (exposure to mediator and mediator to outcome), which will always yield a smaller absolute coefficient compared to its two component coefficients18. Also see Box 3 on power calculations.
Mendelian randomization and the microbiome
In contrast to causal inference, Mendelian randomization is a statistical method from the field of epidemiology, often used to estimate the causal effects of genetic factors on a phenotype in large cohorts.19,20,21 In a nutshell, Mendelian randomization leverages the fact that genotype is fixed at conception and therefore takes place before the manifestation of a phenotype. This, along with other assumptions, allows the researcher to assess causality and directionality of the exposure (genotype) on the outcome (phenotype).
Recently, Mendelian randomization has been applied to microbiome data in the sense that genotype is replaced with microbiome metagenomic content. Particular care is therefore necessary. Unlike genotype, the microbiome is not fixed at conception but remains in constant flux throughout life (Time-varying signals). While it makes sense to assess the causal effect of host genotype on the microbiome, for example, in the case of a host metabolic disorder altering the host gut and hence the microbiome22, it seems much less clear whether taking the microbial metagenome as a fixed exposure is appropriate.
High-dimensional data science
Microbiome–gut–brain axis experiments tend to yield complex, high-dimensional datasets. Here, we will discuss techniques to handle these types of data and strategies to integrate multiple high-dimensional data from the same experiment.
Stratifying and clustering samples
In some cases, it is necessary to stratify data into clusters, distinct subgroups based on microbiome signature. Stratification is a common method of defining enterotypes, which are large subgroups based on microbial taxonomic composition. The precise number of true enterotypes, as well as the best way to define them, is still up for debate (although 3–4 enterotypes are often cited23,24). Initial efforts involved calculating a Jensen–Shannon dissimilarity matrix and performing cluster analysis (clusters here corresponding to enterotypes) using the partition around medoids approach25. More recently, studies have employed the Dirichlet multinomial mixtures approach, a promising technique to estimate enterotypes from the Bayesian school26. In brief, the method involves estimating a probability vector for each sample and then estimating whether these vectors came from the same source (metacommunity, enterotype) or from separate enterotypes. Enterotypes appear to be important constructs because they are related to factors such as host health, diet, and exercise, despite some known limitations27. Notably, bacterial load is not easy to estimate using metagenomic techniques such as 16S and shotgun (although compare ref. 28) but can rather be assessed by pan-bacterial quantitative PCR (qPCR) or, most accurately, using flow cytometry. Bacterial load is associated with enterotype identity and may bias results23,24,27.
Stratifying samples on the basis of feature abundance is a defensible approach under some circumstances, for example, when pursuing functional groups of microorganisms that might exhibit competitive exclusion. However, it is rarely advisable to stratify samples into subgroups while in the middle of an analysis or when working with datasets comprising only 10s–100s of samples because there are too few samples for validation. It is especially important to validate data-driven stratification, either in a new cohort or in a subsection of withheld data that can be used as a validation set. Spurious strata can frequently arise from technical or biological artifacts, leading enthusiastic researchers on long and fruitless tangents. Clustering algorithms, by design, will cluster and can even find seemingly impressive clusters among random noise.
Multi-omics integration
The microbiome refers to the collection of microbial genes in a sample. While the present work focuses on this type of data, other ’omics also exist29,30,31,32. Microbial genetic data provide evidence of microorganism presence as well as their functional potential.33 Besides metagenomics, the three most common types of ’omics data in microbiome–gut–brain axis studies are the following:
-
Metabolomics: the metabolites and small molecules in a sample. Mass spectrometry or nuclear magnetic resonance spectroscopy are the most common techniques to measure the metabolome. Metabolomics can shed light on the functional consequences of a given microbiome.
-
Metatranscriptomics: the sequencing of RNA in a sample. In practice, metatranscriptomics can be thought of as RNAseq on a microbial community rather than a single organism. Metatranscriptomics can tell us about the transcriptional activity of a microbial community. A microorganism may be present and have a certain gene, but it may not be transcribing that gene34.
-
Metaproteomics: the proteins in a sample. Typically, metaproteomics relies on specialized mass spectrometry techniques to identify proteins and derive their sequences. Metaproteomics goes further than metatranscriptomics and tells us whether the transcribed genes are translated to proteins.
There are three broad approaches to data integration, treating datasets as either univariate or multivariate. The suffixes -variable and -variate are often used interchangeably, but they refer to subtly but meaningfully distinct concepts35. In short, -variate refers to the structural nature of the data, whereas -variable refers to the structure and number of variables in the statistical model (also Fig. 2b):
-
Univariate-univariate: with two separate multivariate datasets, one can perform an acceptable analysis using ‘simple’ univariate methods by correlating each microbiome feature (for example, taxa or gene) with individual features in the other dataset, one at a time. Metrics such as Pearson’s and Spearman’s Rank correlation coefficients are commonly used for this purpose. Both of these metrics can be thought of as special cases of a linear model, which we particularly recommend as it allows for the inclusion of covariates.
-
Univariate-multivariate: treat one feature from one dataset as a dependent variable, and use all features from the other dataset as the predictors. By repeating this for each feature, all associations between the datasets are described.
-
Multivariate–multivariate: multivariate regression, such as a canonical correlation analysis or redundancy analysis36, to obtain a single model that associates all features from one dataset with all features from the other dataset. The mixOmics package provides a user-friendly implementation of multivariate methods for microbiome research37,38. Similarly, neural networks or other machine learning can be used39,40,41.
For multivariate–multivariate analysis, one compelling method, DIABLO42, extends this approach by comparing association networks between phenotypes, focusing on the interactions between two ’omics data tests rather than the values within the two individual datasets. This permits the discovery of patterns not necessarily visible in either of the individual datasets. Note also that it is possible to extend any of these approaches to incorporate external information about known relationships within the individual dataset or across the two datasets (for example, via a gene ontology database). For example, joint pathway analysis takes advantage of existing biological knowledge structures by mapping two ’omics datasets to the same metabolic pathways and then assessing the joint coverage as a readout of pathway enrichment43,44. Such knowledge structures could also be leveraged to constrain an analysis to include only feature pairs that are canonically able to interact according to the database, thus potentially preserving power by avoiding unnecessary hypothesis testing, as exemplified by the anansi framework.45 Whatever approach one uses, analysts should take care to normalize or transform their data appropriately, especially since correlations can yield spurious results when measured for compositional data46,47. As with differential abundance analysis, multiple multivariate tests should always be accompanied by an FDR adjustment. When null hypothesis testing is not straightforward in multivariate methods, permutations or algorithmic validation (for example, cross-validation) may be used there instead.
Exploring the mesoscale
Mesoscale features of the microbiome contain information about patterns within parts of a microbiome that can be seen across samples—not necessarily its smallest parts (the microscale) or about the whole system (the macroscale). Mesoscale analysis focuses on identifying community-level patterns that define the ecosystem(s). This is useful because phenomena in a microbiome may be more readily explained by aggregated patterns in the data rather than by any individual feature. The mesoscale is an important object of study in theoretical ecology48. These emerging techniques derive microbiome mesoscale features. The first three make use of external knowledge. The final two are purely data driven:
-
Ecological guilds: ecological guilds are taxonomically unrelated but functionally related clusters of microorganisms that have a shared role in the microbiome (for example, occupy a common niche). For example, microbial communities across a wide span of environments, including soil, the ocean, and the human gut, could be assigned to trophic groups on the basis of how they feed on certain substrates and subsequently pass on metabolites to another trophic group49. While ecological guilds are a promising concept in microbiome science, to our knowledge there are currently no standardized pipelines or databases that can be used to detect and compare ecological guilds across cohorts and experiments (but compare ref. 50). Such tools would be welcome additions to the field51,52.
-
Functional modules: functional modules are a list of curated metabolic pathways encoding for processes that are related to a specific aspect of the microbiome. We will consider two classes of functional modules. Gut–brain modules cover pathways that are related to gut–brain communication, such as serotonin degradation and histamine synthesis. The complete list of gut–brain modules can be accessed as a table in the supplementary files of ref. 53. Gut-–metabolic modules, from the same group, encompass metabolic processes in the microbiome. Changes in gut–metabolic modules can indicate a shift in the microbial metabolic environment and thereby in the fitness landscape, thus allowing for microorganisms with different metabolic features to thrive. The complete list of gut–metabolic modules can be accessed as a table in the supplementary files of the paper that introduced them54. Functional modules are especially interpretable and help develop hypotheses for future experiments. We note that functional module analysis depends on the availability of a functional abundance table (see the section on functions in the companion piece of this Perspective1).
-
Enrichment analysis: differential abundance (DA) analysis is first performed on taxa or genes or other ’omics features, and then a functional database is used to summarize the DA results. In the simplest case, the DA results can be dichotomized into significant or non-significant, and functional status can be dichotomized as present or absent. For each function, one could perform a Fisher exact test (or similar) to test over-enrichment among the significant taxa, genes, or features43,55. Gene set enrichment analysis is a popular generalization of this concept and is commonplace in gene expression analysis55.
-
Network analysis: network analysis is most often applied to study or visualize associations between microbiome features such as taxa or genes. This requires some measure of association. The Pearson’s correlation is the most popular; however, correlations have been shown to yield spurious results when applied to compositional data56. For this reason, several alternatives have been designed specifically for microbiome data46,57,58 (also see the discussion on compositionality in the companion piece to this Perspective1). These metrics build on a log-ratio transformation that makes them more robust to the biases introduced by compositionality59, although they can still be prone to false positives60. A recent benchmark of 213 single-cell datasets has shown that proportionality has excellent performance for sparse high-dimensional data such as those encountered in microbiome research61.
-
Balance selection and summed log-ratios: balance selection and data-driven amalgamation are two new approaches to learning mesoscale features directly from the data. In both cases, the motivation is to find mesoscale features that serve as a biomarker to predict another variable of interest. These mesoscale features are unique in that they are defined explicitly as a ratio between groups of taxa, similar to the Firmicutes-to-Bacteroidetes ratio62. By using a ratio of taxa, any normalization factors would cancel, thus making the method normalization-free. When the groups of taxa are summarized by a geometric mean, the resultant mesoscale feature is called a balance. When they are summarized by a sum, the resultant mesoscale feature is called a summed log-ratio. Software tools such as selbal63, balance64, amalgam65, and CoDaCoRe66 enable analysts to learn mesoscale features in a few lines of code. It is customary to validate the reliability of these features by measuring predictive performance in a withheld test set67.
Time-varying signals
While 16S and shotgun sequencing allow only for snapshot measurements of the microbiome, in reality, microbiomes are dynamic ecosystems in constant flux. To account for this, it has become more common for studies to include multiple (repeated) measures of the microbiome. However, time-series analysis necessitates special considerations68,69.
Statistical considerations with time-varying data analysis
Time-series data, where the same microbiomes are sampled repeatedly, intrinsically break the assumption of independence between samples that many statistical tests rely on. Mixed-effects models are well equipped to handle this type of data, using the resampled microbiome as a random effect. The well-documented and widely used lme4 package in R provides an excellent framework for this70. More-specialized microbiome tools such as MaAsLin271 are also available.
A recent study on the temporal variation of the microbiome estimated that inter-individual variation is smaller than intra-individual variation72. Taking several microbiome measurements over time may therefore be necessary to increase power to detect group differences. Another approach to deal with this high intra-individual variation is to include microbial variance in the model73. This allows investigation of whether microbial variability itself is associated with the phenotype of interest. The idea that microbial variance rather than abundance can be informative for a phenotype is core to the idea of volatility.
Volatility
The microbiome is a dynamic ecosystem that undergoes constant change. The degree of change in the microbiome over time is called volatility, which is inversely related to stability. The term was first coined during the early days of the Human Microbiome Project in the context of instability74 and was soon thereafter used to describe the degree of change in the microbiome between two time points75. It can be helpful to think of volatility as a change in sample diversity (alpha or beta) over time. In a neutral setting, without intervention, a higher volatility is generally considered to be associated with negative health outcomes76. One way to calculate volatility is to measure the beta diversity between two or more time points corresponding to the same host. When measuring volatility in this fashion, it is especially useful to choose a beta diversity metric that is also a distance (that is, follows triangle inequality, such as PhiLR or Aitchison distance) so that any comparisons are standardized for all time points. Volatility has recently been shown to differ between enterotypes, indicating that microbiome composition at least partially explains microbiome volatility72. Because sampling depth is known to affect beta diversity indices, it may be worth subsampling before volatility analysis77.
Circadian rhythms
Circadian rhythms, or 24-hour biological cycles, are key in maintaining physical and mental health78. The microbiome is an example of a biological system that displays such a 24-hour cycle79,80. Typically, models that assess rhythmicity will make use of a sinusoidal model rather than a conventional linear model. Circadian rhythms are a special case of time-varying data as there is an implicit assumption that microbial taxa will oscillate around a set mean (mesor). Due to the 24-hour period of a circadian rhythm, time of sampling becomes an important source of variance and thus a relevant covariate even when the researcher is not interested in investigating circadian rhythms per se. We recently developed the kronos package in R to analyze circadian rhythms in the microbiome81.
Consolidating and looking forward
As the microbiome–gut–brain axis field continues its maturation, we shift our priorities away from a basic demonstration of relevance and toward formulating and addressing more mechanistic questions. In the final section of this Perspective, we briefly look forward to efforts to consolidate findings in the field.
Meta-analyses
In a nutshell, meta-analyses incorporate outcomes from numerous studies on the same subject to estimate a ‘true effect` on the basis of a weighted summary of the component studies. When planning a meta-analysis of microbiome–gut–brain axis studies, it is particularly important to consider which features to analyze. For example, it may be preferable to investigate the role of microbial functions in a disorder rather than taxonomy-level data. In addition, due to large inter-study heterogeneity in methodology, it may not be appropriate to compare reported outcomes from studies at all, and a ‘meta-re-analysis` from raw data may be warranted. This again underlines the importance of making microbiome data publicly available. We note and applaud the burgeoning development of meta-analysis methods for microbiome studies such as MMUPHin by ref. 82, which account for the heterogeneity in pre-processing that precludes standard meta-analysis tools and techniques.83,84,85 A groundswell of attempts at reproducing previous findings and quantitative synthesis of the literature to date will improve the robustness of the field, as it has done in others.
Toward enriching microbiome–gut–brain axis research
In this part 2 of our Bugs as features Perspective, we have taken you on a tour of both adjacent and far-flung topics to enrich contemporary microbiome–gut–brain axis research, from Mendelian randomization and mediation analysis to numerous ways to explore microbiome patterns of the mesoscale. In combination with the concepts and foundations detailed in part 1, and the corresponding supplementary code tutorial, we have described the key considerations for microbiome–gut–brain axis analysis1. In our opinion, establishing causality, integrating multi-omics data, and accounting for the dynamic nature of the microbiome are key. We hope that this Perspective has assisted with confident navigation of the microbial landscape. We trust that the increased use of biologically and statistically sound methods such as those described here will improve our understanding of the complex phenomenon known as the microbiome–gut–brain axis.
References
Bastiaanssen, T. F. S., Quinn, T. P. & Loughman, A. Bugs as features (part 1): concepts and foundations for the compositional data analysis of the microbiome–gut–brain axis. Nat. Ment. Health https://doi.org/10.1038/s44220-023-00148-3 (2023).
Walter, J., Armet, A. M., Finlay, B. B. & Shanahan, F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232 (2020).
Bastiaanssen, T. F. S. & Cryan, J. F. The microbiota–gut–brain axis in mental health and medication response: parsing directionality and causality. Int. J. Neuropsychopharmacol. 24, 216–220 (2021).
Sugihara, G. et al. Detecting causality in complex ecosystems. Science 338, 496–500 (2012).
Koch, R. Untersuchungen uber bakterien v. die aetiologie der milzbrand-krankheit, begrunder auf die entwicklungegeschichte bacillus anthracis. Beitrage zur biologie der Pflanzen 2, 277–310 (1877).
Hill, A. B. The environment and disease: association or causation? Proc. R. Soc. Med. 58, 295–300 (1965).
Hernán, M. A. The c-word: scientific euphemisms do not improve causal inference from observational data. Am. J. Public Health 108, 616–619 (2018).
VanderWeele, T. J. & Robins, J. M. Directed acyclic graphs, sufficient causes, and the properties of conditioning on a common effect. Am. J. Epidemiol. 166, 1096–1104 (2007).
Zhu, F. et al. Metagenome-wide association of gut microbiome features for schizophrenia. Nat. Commun. 11, 1612 (2020).
MacKinnon, D. P., Fairchild, A. J. & Fritz, M. S. Mediation analysis. Annu. Rev. Psychol. 58, 593–614 (2007).
Logan, A. C. & Jacka, F. N. Nutritional psychiatry research: an emerging discipline and its intersection with global urbanization, environmental challenges and the evolutionary mismatch. J. Physiol. Anthropol. https://doi.org/10.1186/1880-6805-33-22 (2014).
Cryan, J. F. et al. The microbiota–gut–brain axis. Physiol. Rev. 99, 1877-2013 (2019).
Yap, C. X. et al. Autism-related dietary preferences mediate autism–gut microbiome associations. Cell 184, 5916–5931 (2021).
Morton, J. T., Donovan, S. M. & Taroncher-Oldenburg, G. Decoupling diet from microbiome dynamics results in model mis-specification that implicitly annuls potential associations between the microbiome and disease phenotypes—ruling out any role of the microbiome in autism (Yap et al. 2021) likely a premature conclusion. Preprint at bioRxiv https://doi.org/10.1101/2022.02.25.482051 (2022).
Tingley, D., Yamamoto, T., Hirose, K., Keele, L. & Imai, K. mediation: R package for causal mediation analysis. J. Stat. Softw. https://doi.org/10.18637/jss.v059.i05 (2014).
Fairchild, A. J. & McDaniel, H. L. Best (but oft-forgotten) practices: mediation analysis. Am. J. Clin. Nutr. 105, 1259–1271 (2017).
Cain, M. K., Zhang, Z. & Bergeman, C. Time and other considerations in mediation design. Educ. Psychol. Meas. 78, 952–972 (2018).
Baron, R. M. & Kenny, D. A. The moderator–mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J. Pers. Soc. Psychol. 51, 1173 (1986).
Smith, G. D. & Ebrahim, S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22 (2003).
Gagliano Taliun, S. A. & Evans, D. M. Ten simple rules for conducting a Mendelian randomization study. PLoS Comput. Biol. 17, e1009238 (2021).
Sanderson, E. et al. Mendelian randomization. Nat. Rev. Methods Primers 2, 6 (2022).
Sanna, S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605 (2019).
Costea, P. I. et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16 (2018).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Holmes, I., Harris, K. & Quince, C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE 7, e30126 (2012).
Knights, D. et al. Rethinking ‘enterotypes’. Cell Host Microbe 16, 433–437 (2014).
Cruz, G. N. F., Christoff, A. P. & de Oliveira, L. F. V. Equivolumetric protocol generates library sizes proportional to total microbial load in 16S amplicon sequencing. Front. Microbiol. 12, 425 (2021).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Smolinska, A. et al. Volatile metabolites in breath strongly correlate with gut microbiome in CD patients. Anal. Chim. Acta 1025, 1–11 (2018).
Tang, Z.-Z. et al. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front.Genet. 10, 00454 (2019).
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976 (2019).
Aguiar-Pulido, V. et al. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis: supplementary issue: bioinformatics methods and applications for big metagenomics data. Evol. Bioinform. 12, EBO–S36436 (2016).
Abu-Ali, G. S. et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat. Microbiol. 3, 356–366 (2018).
Mallick, H. et al. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 18, 228 (2017).
Meng, C. et al. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinform. 17, 628–641 (2016).
Lê Cao, K.-A., Rossouw, D., Robert-Granié, C. & Besse, P. A sparse PLS for variable selection when integrating omics data. Stat. Appl. Genet. Mol. Biol. 7 35 (2008).
Rohart, F., Gautier, B., Singh, A. & Lê Cao, K.-A. mixomics: an r package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752 (2017).
Le, V., Quinn, T. P., Tran, T. & Venkatesh, S. Deep in the bowel: highly interpretable neural encoder–decoder networks predict gut metabolites from gut microbiome. BMC Genom. 21, 256 (2020).
Morton, J. T. et al. Learning representations of microbe–metabolite interactions. Nat. Methods 16, 1306–1314 (2019).
Reiman, D., Layden, B. T. & Dai, Y. Mimenet: exploring microbiome–metabolome relationships using neural networks. PLoS Comput. Biol. 17, e1009021 (2021).
Singh, A. et al. Diablo: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 35, 3055–3062 (2019).
Chong, J. et al. Metaboanalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018).
Pang, Z. et al. Metaboanalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396 (2021).
Bastiaanssen, T. F. S., Quinn, T. P. & Cryan, J. F. Knowledge-based integration of multi-omic datasets with anansi: annotation-based analysis of specific interactions. Preprint at https://arxiv.org/abs/2305.10832 (2023).
Quinn, T. P., Richardson, M. F., Lovell, D. & Crowley, T. M. propr: an r-package for identifying proportionally abundant features using compositional data analysis. Sci. Rep. 7, 16252 (2017).
Quinn, T. P. & Erb, I. Examining microbe–metabolite correlations by linear methods. Nat. Methods 18, 37–39 (2021).
Hogeweg, P. in Simulating Complex Systems by Cellular Automata (eds Kroc, J. et al.) 19–28 (Springer, 2010).
Gralka, M., Szabo, R., Stocker, R. & Cordero, O. X. Trophic interactions and the drivers of microbial community assembly. Curr. Biol. 30, R1176–R1188 (2020).
Frioux, C. et al. Enterosignatures define common bacterial guilds in the human gut microbiome. Cell Host Microbe 31, 1111–1125 (2023).
Lam, Y. Y., Zhang, C. & Zhao, L. Causality in dietary interventions—building a case for gut microbiota. Genome Med. 10, 62 (2018).
Zhao, L. et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018).
Valles-Colomer, M. et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 4, 623–632 (2019).
Vieira-Silva, S. et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 1, 16088 (2016).
Irizarry, R. A., Wang, C., Zhou, Y. & Speed, T. P. Gene set enrichment analysis made simple. Stat. Methods Med. Res. 18, 565–575 (2009).
Lovell, D., Pawlowsky-Glahn, V., Egozcue, J. J., Marguerat, S. & Bähler, J. Proportionality: a valid alternative to correlation for relative data. PLoS Comput. Biol. 11, e1004075 (2015).
Friedman, J. & Alm, E. J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8, e1002687 (2012).
Kurtz, Z. D. et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11, e1004226 (2015).
Quinn, T. P., Erb, I., Richardson, M. F. & Crowley, T. M. Understanding sequencing data as compositions: an outlook and review. Bioinformatics 34, 2870–2878 (2018).
Erb, I. & Notredame, C. How should we measure proportionality on relative gene expression data? Theory Biosci. 135, 21–36 (2016).
Skinnider, M. A., Squair, J. W. & Foster, L. J. Evaluating measures of association for single-cell transcriptomics. Nat. Methods 16, 381–386 (2019).
Mariat, D. et al. The firmicutes/bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9, 123 (2009).
Rivera-Pinto, J. et al. Balances: a new perspective for microbiome analysis. mSystems 3, e00053-18 (2018).
Quinn, T. P. Visualizing balances of compositional data: a new alternative to balance dendrograms. F1000Res. 7, 1278 (2018).
Quinn, T. P. & Erb, I. Amalgams: data-driven amalgamation for the dimensionality reduction of compositional data. NAR Genom. Bioinform. 2, lqaa076 (2020).
Gordon-Rodriguez, E., Quinn, T. P. & Cunningham, J. P. Learning sparse log-ratios for high-throughput sequencing data. Bioinformatics 38, 157–163 (2021).
Quinn, T. P., Gordon-Rodriguez, E. & Erb, I. A critique of differential abundance analysis, and advocacy for an alternative. Preprint at https://arxiv.org/abs/2104.07266 (2021).
Kodikara, S., Ellul, S. & Lê Cao, K.-A. Statistical challenges in longitudinal microbiome data analysis. Brief. Bioinform. 23, bbac273 (2022).
Bokulich, N. A. et al. q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems 3, e00219-18 (2018).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. https://doi.org/10.18637/jss.v067.i01 (2015).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Vandeputte, D. et al. Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat. Commun. 12, 6740 (2021).
Martino, C. et al. Context-aware dimensionality reduction deconvolutes gut microbial community dynamics. Nat. Biotechnol. 39, 165–168 (2021).
Weinstock, G. M. The volatile microbiome. Genome Biol. 12, 114 (2011).
Goodrich, J. K. et al. Conducting a microbiome study. Cell 158, 250–262 (2014).
Bastiaanssen, T. F. S. et al. Volatility as a concept to understand the impact of stress on the microbiome. Psychoneuroendocrinology 124, 105047 (2021).
Park, D. J. & Plantinga, A. M. Impact of data and study characteristics on microbiome volatility estimates. Genes 14, 218 (2023).
Caliyurt, O. Role of chronobiology as a transdisciplinary field of research: its applications in treating mood disorders. Balkan Med. J. 34, 514–521 (2017).
Thaiss, C. A. et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014).
Liang, X., Bushman, F. D. & FitzGerald, G. A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl Acad. Sci. USA 112, 10479–10484 (2015).
Bastiaanssen, T. F. S. et al. Kronos: a computational tool to facilitate biological rhythmicity analysis. Preprint at bioRxiv https://doi.org/10.1101/2023.04.21.537503 (2023).
Ma, S. et al. Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using mmuphin. Genome Biol. 23, 208 (2022).
Duvallet, C., Gibbons, S. M., Gurry, T., Irizarry, R. A. & Alm, E. J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8, 1784 (2017).
Chong, J., Liu, P., Zhou, G. & Xia, J. Using microbiomeanalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821 (2020).
Morton, J. T. et al. Multi-level analysis of the gut–brain axis shows autism spectrum disorder-associated molecular and microbial profiles. Nat. Neurosci. 26, 1208–1217 (2023).
Wortelboer, K., Nieuwdorp, M. & Herrema, H. Fecal microbiota transplantation beyond clostridioides difficile infections. EBioMedicine 44, 716–729 (2019).
Kelly, J. R. et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 82, 109–118 (2016).
Boehme, M. et al. Microbiota from young mice counteracts selective age-associated behavioral deficits. Nat. Aging 1, 666–676 (2021).
Gheorghe, C. E. et al. Investigating causality with fecal microbiota transplantation in rodents: applications, recommendations and pitfalls. Gut Microbes 13, 1941711 (2021).
Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145 (2018).
Podlesny, D. et al. Metagenomic strain detection with samestr: identification of a persisting core gut microbiota transferable by fecal transplantation. Microbiome 10, 53 (2022).
Valles-Colomer, M. et al. Variation and transmission of the human gut microbiota across multiple familial generations. Nat. Microbiol. 7, 87–96 (2022).
Valles-Colomer, M. et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614, 125–135 (2023).
Johnson, J. S. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029 (2019).
Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01688-w (2023).
Ponsonby, A.-L. Reflection on modern methods: building causal evidence within high-dimensional molecular epidemiological studies of moderate size Int. J. Epidemiol. 50, 1016–1029 (2021).
VanderWeele, T. J., Hernán, M. A. & Robins, J. M. Causal directed acyclic graphs and the direction of unmeasured confounding bias. Epidemiology 19, 720–728 (2008).
VanderWeele, T. J. Principles of confounder selection. Eur. J. Epidemiol. 34, 211–219 (2019).
McLaren, M. R., Willis, A. D. & Callahan, B. J. Consistent and correctable bias in metagenomic sequencing experiments. Elife 8, e46923 (2019).
Wang, Y. & LêCao, K.-A. Managing batch effects in microbiome data. Brief Bioinform. 21, 1954–1970 (2020).
Schisterman, E. F., Cole, S. R. & Platt, R. W. Overadjustment bias and unnecessary adjustment in epidemiologic studies. Epidemiology 20, 488–495 (2009).
Textor, J., van der Zander, B., Gilthorpe, M. S., Liśkiewicz, M. & Ellison, G. T. Robust causal inference using directed acyclic graphs: the r package ‘dagitty’. Int. J. Epidemiol. 45, 1887–1894 (2016).
Bross, I. D. Spurious effects from an extraneous variable. J. Chronic Dis. 19, 637–647 (1966).
Hernán, M. Causal diagrams: draw your assumptions before your conclusions; https://pll.harvard.edu/course/causal-diagrams-draw-your-assumptions-your-conclusions (Harvard Univ., 2023).
Hernán, M. A. & Robins, J. A. Causal Inference: What If (Chapman & Hall/CRC, 2020).
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 (2003).
Acknowledgments
We thank A.-L. Ponsonby for her expert comments on DAGs, D. L. Dahly for his insights on statistical analysis, and J. F. Cryan for his excellent advice. We are grateful for their help and support. APC Microbiome Ireland is a research center funded by Science Foundation Ireland (SFI), through the Irish Governments’ national development plan (grant no. 12/RC/2273_P2).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Mental Health thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bastiaanssen, T.F.S., Quinn, T.P. & Loughman, A. Bugs as features (part 2): a perspective on enriching microbiome–gut–brain axis analyses. Nat. Mental Health 1, 939–949 (2023). https://doi.org/10.1038/s44220-023-00149-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44220-023-00149-2
This article is cited by
-
The gut virome is associated with stress-induced changes in behaviour and immune responses in mice
Nature Microbiology (2024)