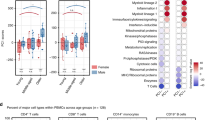
Abstract
Frailty is an intermediate status of the human aging process, associated with decompensated homeostasis and death. The immune phenotype of frailty and its underlying cellular and molecular processes remain poorly understood. We profiled 114,467 immune cells from cord blood, young adults and healthy and frail old adults using single-cell RNA and TCR sequencing. Here we show an age-dependent accumulation of transcriptome heterogeneity and variability in immune cells. Characteristic transcription factors were identified in given cell types of specific age groups. Trajectory analysis revealed cells from non-frail and frail old adults often fall into distinct paths. Numerous TCR clonotypes were shared among T-cell subtypes in old adults, indicating differential pluripotency and resilience capabilities of aged T cells. A frailty-specific monocyte subset was identified with exclusively high expression of long noncoding RNAs NEAT1 and MALAT1. Our study discovers human frailty-specific immune cell characteristics based on the comprehensive dimensions in the immune landscape of aging and frailty.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Single-cell sequencing data presented in this paper are deposited in the Gene Expression Omnibus data repository under accession code GSE157007. Human genome reference GRCh38 (v.1.2.0) was downloaded from https://support.10xgenomics.com/single-cell-gene-expression/software/release-notes/build/.
Code availability
Analytical scripts and codes are available upon request from the corresponding authors.
References
Reynolds, L. M. et al. Transcriptomic profiles of aging in purified human immune cells. BMC Genomics 16, 333 (2015).
Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570 (2015).
Vaupel, J. M., Manton, K. G. & Stallard, E. The impact of heterogeneity in individual frailty on the dynamics of mortality. Demography 16, 439–454 (1979).
Dent, E. et al. The Asia–Pacific Clinical Practice Guidelines for the Management of Frailty. J. Am. Med. Dir. Assoc. 18, 564–575 (2017).
Morley, J. E. et al. Frailty consensus: a call to action. J. Am. Med. Dir. Assoc. 14, 392–397 (2013).
Wilson, D., Jackson, T., Sapey, E. & Lord, J. M. Frailty and sarcopenia: the potential role of an aged immune system. Ageing Res. Rev. 36, 1–10 (2017).
Lei, W. et al. Applications of high-throughput ‘omics’ data in the study of frailty. Transl. Med. Aging 3, 40–51 (2019).
Merkerova, M. et al. Differential gene expression in umbilical cord blood and maternal peripheral blood. Eur. J. Haematol. 83, 183–190 (2009).
Uhlen, M. et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366, eaax9198 (2019).
Muller, L., Di Benedetto, S. & Pawelec, G. The immune system and its dysregulation with aging. Subcell. Biochem. 91, 21–43 (2019).
Nikolich-Zugich, J. The twilight of immunity: emerging concepts in aging of the immune system. Nat. Immunol. 19, 10–19 (2018).
Britanova, O. V. et al. Dynamics of individual T cell repertoires: from cord blood to centenarians. J. Immunol. 196, 5005–5013 (2016).
Krishna, C., Chowell, D., Gonen, M., Elhanati, Y. & Chan, T. A. Genetic and environmental determinants of human TCR repertoire diversity. Immun. Ageing 17, 26 (2020).
Bagaev, D. V. et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 48, D1057–D1062 (2020).
Kimmel, J. C. et al. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 29, 2088–2103 (2019).
Enge, M. et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330 (2017).
Martinez-Jimenez, C. P. et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355, 1433–1436 (2017).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
McConnell, B. B. & Yang, V. W. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 90, 1337–1381 (2010).
Ginsberg, M. et al. Amino acid residues required for physical and cooperative transcriptional interaction of STAT3 and AP-1 proteins c-Jun and c-Fos. Mol. Cell. Biol. 27, 6300–6308 (2007).
Bejjani, F., Evanno, E., Zibara, K., Piechaczyk, M. & Jariel-Encontre, I. The AP-1 transcriptional complex: local switch or remote command? Biochim. Biophys. Acta Rev. Cancer 1872, 11–23 (2019).
Zhao, J. et al. TLR2 involved in naive CD4+ T cells rescues stress-induced immune suppression by regulating TH1/TH2 and TH17. Neuroimmunomodulation 22, 328–336 (2015).
Dozmorov, M. G., Coit, P., Maksimowicz-McKinnon, K. & Sawalha, A. H. Age-associated DNA methylation changes in naive CD4+ T cells suggest an evolving autoimmune epigenotype in aging T cells. Epigenomics 9, 429–445 (2017).
Donnini, A. et al. Age-related susceptibility of naive and memory CD4 T cells to apoptosis induced by IL-2 deprivation or PHA addition. Biogerontology 6, 193–204 (2005).
Gupta, S., Su, H., Agrawal, S. & Gollapudi, S. Molecular changes associated with increased TNF-alpha-induced apoptosis in naive (TN) and central memory (TCM) CD8+ T cells in aged humans. Immun. Ageing 15, 2 (2018).
Kunstle, G. et al. Identification of Akt association and oligomerization domains of the Akt kinase coactivator TCL1. Mol. Cell. Biol. 22, 1513–1525 (2002).
Pei, L., Castrillo, A. & Tontonoz, P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol. Endocrinol. 20, 786–794 (2006).
Merino, A. M., Kim, H., Miller, J. S. & Cichocki, F. Unraveling exhaustion in adaptive and conventional NK cells. J. Leukoc. Biol. 108, 1361–1368 (2020).
Aalijahan, H. & Ghorbian, S. Long non-coding RNAs and cervical cancer. Exp. Mol. Pathol. 106, 7–16 (2019).
West, J. A. et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 (2014).
Zhang, F. et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 75, 96–104 (2016).
Zhang, P., Cao, L., Zhou, R., Yang, X. & Wu, M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 10, 1495 (2019).
Zheng, Y. et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell 11, 740–770 (2020).
Huang, Z. et al. Effects of sex and aging on the immune cell landscape as assessed by single-cell transcriptomic analysis. Proc. Natl Acad. Sci. USA 118, e2023216118 (2021).
Aging Atlas Consortium. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 49, D825–D830 (2021).
Hashimoto, K. et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4+ T cells in supercentenarians. Proc. Natl Acad. Sci. USA 116, 24242–24251 (2019).
Chen, G. et al. Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens. Cell Rep. 19, 569–583 (2017).
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018).
Butler, A. A., Johnston, D. R., Kaur, S. & Lubin, F. D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 12, eaaw9277 (2019).
Hamann, P. D. et al. Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis. Sci. Rep. 7, 8024 (2017).
Yang, H. et al. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget 8, 77400–77406 (2017).
Mitnitski, A. B., Mogilner, A. J. & Rockwood, K. Accumulation of deficits as a proxy measure of aging. ScientificWorldJournal 1, 323–336 (2001).
Searle, S. D., Mitnitski, A., Gahbauer, E. A., Gill, T. M. & Rockwood, K. A standard procedure for creating a frailty index. BMC Geriatr. 8, 24 (2008).
Yin, J. H., Zeng, Y. B., Zhou, Z. & Fang, Y. Study on the status of frailty and related determinants among the elderly in China. Zhonghua Liu Xing Bing Xue Za Zhi 39, 1244–1248 (2018).
Yamazaki, T., Fujikawa, C., Kubota, A., Takahashi, A. & Hirose, T. CRISPRa-mediated NEAT1 lncRNA upregulation induces formation of intact paraspeckles. Biochem. Biophys. Res. Commun. 504, 218–224 (2018).
Stuart, T. et al. Comprehensive Integration of single-cell data. Cell 177, 1888–1902 (2019).
Kolodziejczyk, A. A. et al. Single-cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17, 471–485 (2015).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Acknowledgements
This work was supported by funding from the National Key Research and Development Program of China (2021YFC2009404 to G.Z., 2018YFC2002003 to G.C., 2018YFC2002001 to J.G., 2018YFC2002002 to J.C., 2018YFC2002004 to N.P. and 2018YFC2002000 to Z.B.), the Natural Science Foundation of China (81971301 and 32050410285 to O.J.L., U1801285 to G.C., 81973221 to W.G. and 81971327 to E.F.F.), the Guangzhou Planned Project of Science and Technology (202002020039 to O.J.L., 202102010030 to G.Z. and 201904010111 to G.C.), the Initial Supporting Foundation of Jinan University (to O.J.L. and G.C.), the Milstein Medical Asian American Partnership (MMAAP) Foundation of the USA under the Irma and Paul Milstein Program for Senior Health (to G.C. and S.X.L.), the China Pharmaceutical University 2020 Innovation and Entrepreneurship Elite Cultivation Plan (to W.G.), Helse Sør-Øst (2017056 and 2020001 to E.F.F.) and the Research Council of Norway (262175 and 277813 to E.F.F.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank R. Williams for critical proofreading of the paper and suggestions for improvements.
Author information
Authors and Affiliations
Contributions
O.J.L., F.L. and G.C. conceived and designed this study. F.L. and H.L. provided clinical samples. G.Z., J.C., Z.L. and J.S. prepared the samples. W.L. performed cell sorting and experiments. Y.X., C.X. and L.G. helped with the experiments. O.J.L., R.Z. and G.C. performed data analysis. L.T. and W.G. built the antigen-specific TCR database. J.G., N.P., J.C. and Z.B. helped with clinical information analysis. H.S., Z.-J.Z., E.F.F., Y.R., S.X.L., Z.J. and Z.B. helped with data analysis and interpretation. O.J.L., W.L. and G.C. wrote the manuscript with input from Y.R.
Corresponding authors
Ethics declarations
Competing interests
E.F.F. has a CRADA arrangement with ChromaDex and is a consultant to Aladdin Healthcare Technologies and the Vancouver Dementia Prevention Centre. The other authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Summary of samples, scRNA-Seq data quality assessment, selected marker gene expression, cell surface protein expression, productive TCR detection projected on single cells, Cord blood-specific gene expression and cell type composition by individual sample.
(A) Schematic of single-cell transcriptome, TCR and cell surface protein profiling of mononuclear cells (MCs) from cord blood and peripheral blood of Young, Old and Frail donors. (B) Summary of chronological age and grouping of samples. (C) Violin plots and boxplots of number of unique genes (left) and percentage of mitochondria genes (right) detected by scRNA-Seq data in each sample. The horizontal dashed lines in the left panel mark 200 and 3,000, respectively. The horizontal dashed line in the right panel marks 10%. Mt: mitochondria. The number of retained cells per sample is shown in Fig. 1a. (D) PCA visualization of scRNA-Seq data split by age groups. The PCA plots showed no obvious age group bias nor batch effect in scRNA-Seq data. (E) Projection of expression levels of selected marker genes on UMAP visualization. (F) Expression levels of cell surface proteins projected onto the same UMAP visualization as Fig. 1b. (G) Productive TCR detection projected onto UMAP. (H) Violin plots of IFIT2, IFIT3, PRDX2 and SOX4 expression in cord blood samples across all cell types (x-axis). Refer to Fig. 1b for cell type numbering. IFIT2 and IFIT3 are adult-specific genes, and PRDX2 and SOX4 are fetus-specific genes. (I) Scatter plots of T cell, B cell, NK cell and monocyte proportion in each donor. The blue lines were fitted by linear model excluding the Cord blood samples. Shaded areas around the fitted lines indicate the confidence intervals. Pearson’s correlations for the fitted samples are shown. Stack plot of proportion of cell types across each donor. From left to right, the order of the donors is the same as c. Refer to Fig. 1b for cell type numbering.
Extended Data Fig. 2 Cell surface protein expression and TCR detection of T cells.
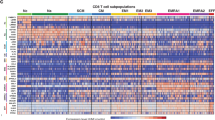
(A) Expression levels of selected marker genes projected onto the same UMAP visualization as Fig. 1f. (B) Detection levels of cell surface proteins projected onto the same UMAP visualization as Fig. 1f. (C) Boxplots of cell surface protein CCR7 and CD45RA (left), and CD4 and CD8 (right) expression in distinct T cell subtypes. *: P < 0.05 (Wilcoxon Rank Sum test, two-sided). The exact cell number per cell type is summarized in Supplementary Table 4. (D) Detection of productive TCR sequences by age groups projected onto UMAP visualization. Cluster 1-6 refers to Naïve T, CD4 Tcm, CD8 Tcm, CD8+ Tem, Treg and Exhausted T cells, respectively. (E) T cell subtype-shared TCR clonotypes projected onto UMAP visualization and split by individual sample. Only shared clonotypes with >100 copies are shown.
Extended Data Fig. 3 Gene expression variance comparison across age groups and additional examples of age group specific genes visualized as violin plots.
(A) Summary of average gene expression overdispersion by cell type and age group. Higher value indicates genes in the corresponding cell type of an age group were expressed with a wider spectrum, and vice versa. Data are presented as mean±SEM. *: P < 0.05; **: P < 0.01; ***: P < 0.001 (One-way ordinary ANOVA). The exact cell number per cell type is summarized in Supplementary Table 4. (B) Violin plots of expression levels of additional age group specific genes in all cell types. From left to right, the order of cell types is the same as a. * denotes the expression of the appointed group is significantly (FDR < 0.05; Wilcoxon Rank Sum test, two-sided with Benjamini-Hochberg adjustment) different from the other three groups combined.
Extended Data Fig. 4 Age-group/cell-type specific expression regulatory programs in Cord blood and Young, and supportive data for Fig. 4.
a. Heatmap visualization of average AUC scores (regulon activities) for cells that are of specific regulon activity in at least one age-group/cell-type combination. The corresponding TFs for the regulons are shown on the right. ‘extended’ indicates the motifs of the TFs are enriched in the ±10 kb regions around the TSSs of the corresponding target genes. b. UMAP visualization of expression levels and AUC scores of RELB in B cells and monocytes across the age groups. c. Density plot of the number of target genes in regulons. The vertical line marks 100 genes. Regulons with ≤100 genes are considered as non-ubiquitous targeting regulons by the corresponding TF, otherwise ubiquitous. d-e. Cord blood (d) and Young (e) group-specific regulons visualized as networks. Color and shape coding are the same as Fig. 4b, c. f-g. Boxplots of mean expression levels of target genes in Cord blood (f) and Young (g) group-specific regulons. Corresponding TFs are shown underneath each boxplot. # indicates the binding motifs of the TF are enriched in the ±10 kb regions around the TSSs of target genes, otherwise ±500 bp. * indicates the mean expression values in the appointed group is significantly different (FDR < 0.05; Wilcoxon Rank Sum test, two-sided with Benjamini-Hochberg adjustment) from the other three groups combined. h. Boxplots of MAFB and selected MAFB target genes expression in monocytes across the age groups. *: FDR < 0.05 (Wilcoxon Rank Sum test, two-sided with Benjamini-Hochberg adjustment). In f, g and h, the exact cell number per cell type is summarized in Supplementary Table 4.
Extended Data Fig. 5 Immune-ageing trajectory of CD8 Tem cells.
a-b. DDR (discriminative dimensionality reduction) tree visualization of CD8 Tem cell trajectory with pseudotime (a) and age group (b) information mapped on. c. Top: Heatmap visualization of expression levels of genes with correlated and anti-correlated expression profile to CD8 Tem cell immune-ageing pseudotime. Top: The x axis represents pseudotime ordering. Gene expression levels across the pseudotime are z-score transformed and smoothed. Bottom: Age group annotation of cells aligned along the pseudotime axis. The number of cells from each age group is labelled on the right.
Extended Data Fig. 6 Expression of selected NK cell specific genes projected onto UMAP visualization.
a. Expression levels of selected NK cell specific genes projected onto the same UMAP as Fig. 1b. NK 1 and 2 (cluster 5 and 8 in Fig. 1b) both exhibited high expression in NKG7, GNLY and GZMB. HLA-DRB1, HLA-DPA1 and HLA-DPB1 had higher expression towards the NK 1 side (cluster 5); CD69, NFKBIZ and FOS had higher expression on the NK 2 side (cluster 8).
Extended Data Fig. 7 Monocyte subtype characterization and gender-specific gene expression for Old and Frail age group across all cell types.
a. UMAP visualization of subtypes of monocytes by single-cell transcriptome profiles. The cells reanalyzed and shown here consists of the cells in cluster 4, 10 and 12 of the original clustering results (Fig. 1b). b. Expression levels of genes specific to different types of monocytes projected onto the same UMAP visualization as a. c. Variation of proportion of each identified monocyte subtype across age groups. See also Supplementary Table 4. d. Heatmap visualization of gender-specific gene expression for Old and Frail age group. Gender-specific genes were identified by comparing the whole-transcriptome profiles between matching number of female and male donors with similar frail index for each cell type. Only genes significantly differentially expressed (FDR < 0.05; Wilcoxon Rank Sum test with Benjamini-Hochberg adjustment) in at least 5 cell types are shown. Genes from chrX and chrY are highlighted with red and blue dots, respectively. The order of each cell type (column) is the same as Supplementary Fig. 3a.
Supplementary information
Rights and permissions
About this article
Cite this article
Luo, O.J., Lei, W., Zhu, G. et al. Multidimensional single-cell analysis of human peripheral blood reveals characteristic features of the immune system landscape in aging and frailty. Nat Aging 2, 348–364 (2022). https://doi.org/10.1038/s43587-022-00198-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-022-00198-9
This article is cited by
-
Senescent immune cells accumulation promotes brown adipose tissue dysfunction during aging
Nature Communications (2023)
-
scRNA-seq revealed the special TCR β & α V(D)J allelic inclusion rearrangement and the high proportion dual (or more) TCR-expressing cells
Cell Death & Disease (2023)
-
scBCR-seq revealed a special and novel IG H&L V(D)J allelic inclusion rearrangement and the high proportion dual BCR expressing B cells
Cellular and Molecular Life Sciences (2023)
-
Insufficient epitope-specific T cell clones are responsible for impaired cellular immunity to inactivated SARS-CoV-2 vaccine in older adults
Nature Aging (2023)
-
Immune-Ageing Evaluation of Peripheral T and NK Lymphocyte Subsets in Chinese Healthy Adults
Phenomics (2023)