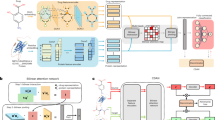
Abstract
Conditional domain adversarial learning presents a promising approach for enhancing the generalizability of deep learning-based methods. Inspired by the efficacy of conditional domain adversarial networks, Bai and colleagues introduced DrugBAN, a methodology designed to explicitly learn pairwise local interactions between drugs and targets. DrugBAN leverages drug molecular graphs and target protein sequences, employing conditional domain adversarial networks to improve the ability to adapt to out-of-distribution data and thereby ensuring superior prediction accuracy for new drug–target pairs. Here we examine the reusability of DrugBAN and extend the evaluation of its generalizability across a wider range of biomedical contexts beyond the original datasets. Various clustering-based strategies are implemented to resplit the source and target domains to assess the robustness of DrugBAN. We also apply this cross-domain adaptation technique to the prediction of cell line–drug responses and mutation–drug associations. The analysis serves as a stepping-off point to better understand and establish a general template applicable to link prediction tasks in biomedical bipartite networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data used in our study are available via GitHub at https://github.com/vshy-dream/DrugBAN-reusability (ref. 22). All of the data used in the original paper by Bai et al. for testing DrugBAN are available via GitHub at https://github.com/peizhenbai/DrugBAN/tree/main/datasets (ref. 23).
Code availability
The original DrugBAN code is available via GitHub at https://github.com/peizhenbai/DrugBAN (ref. 23). Our modified DrugBAN version with changes is available via GitHub at https://github.com/vshy-dream/DrugBAN-reusability (ref. 22).
References
Gosak, M. et al. Network science of biological systems at different scales: a review. Phys. Life Rev. 24, 118–135 (2018).
Su, C., Tong, J., Zhu, Y., Cui, P. & Wang, F. Network embedding in biomedical data science. Brief. Bioinform. 21, 182–197 (2020).
Abbasi, K. et al. DeepCDA: deep cross-domain compound–protein affinity prediction through LSTM and convolutional neural networks. Bioinformatics 36, 4633–4642 (2020).
Bai, P., Miljković, F., John, B. & Lu, H. Interpretable bilinear attention network with domain adaptation improves drug–target prediction. Nat. Mach. Intell. 5, 126–136 (2023).
Long, M., Cao, Z., Wang, J. & Jordan, M. I. Conditional adversarial domain adaptation. In Advances in Neural Information Processing Systems, Curran Associates, Inc. 31, 1647–1657 (2018).
Chen, L. et al. TransformerCPI: improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36, 4406–4414 (2020).
Liu, T., Lin, Y., Wen, X., Jorissen, R. N. & Gilson, M. K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 35, D198–D201 (2007).
Zitnik, M., Sosič, R., Maheshwari, S., & Leskovec, J. BioSNAP datasets: Stanford biomedical network dataset collection, https://snap.stanford.edu/biodata/ (2018).
Hartigan, J. A. & Wong, M. A. Algorithm AS 136: a K-means clustering algorithm. J. R. Stat. Soc. Ser. C 28, 100–108 (1979).
Davis, M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Tang, J. et al. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J. Chem. Inf. Model. 54, 735–743 (2014).
Wu, H. et al. AttentionMGT-DTA: a multi-modal drug-target affinity prediction using graph transformer and attention mechanism. Neur. Netw. 169, 623–636 (2024).
Caldwell, G. W., Yan, Z., Lang, W. & Masucci, J. A. The IC50 concept revisited. Curr. Top. Med. Chem. 12, 1282–1290 (2012).
Fu, H., Huang, F., Liu, X., Qiu, Y. & Zhang, W. MVGCN: data integration through multi-view graph convolutional network for predicting links in biomedical bipartite networks. Bioinformatics 38, 426–434 (2022).
Murtagh, F. & Contreras, P. Algorithms for hierarchical clustering: an overview. WIREs Data Min. Knowl. Discov. 2, 86–97 (2012).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Yang, W. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucl. Acids Res. 41, D955–D961 (2013).
Feuk, L., Carson, A. & Scherer, S. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 (2006).
Sondka, Z. et al. COSMIC: a curated database of somatic variants and clinical data for cancer. Nucl. Acids Res. 52, D1210–D1217 (2024).
Wagner, A. H. et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 52, 448–457 (2020).
Nikam, R. & Gromiha, M. M. Seq2Feature: a comprehensive web-based feature extraction tool. Bioinformatics 35, 4797–4799 (2019).
Xu, T. et al. vshy-dream/DrugBAN-reusability. Zenodo https://doi.org/10.5281/zenodo.10780639 (2024).
Bai, P. & Lu, H. peizhenbai/DrugBAN. GitHub https://github.com/peizhenbai/DrugBAN (2023).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number 62102004) and the Introduction and Stabilization of Talent Project of Anhui Agricultural University (grant number yj2019-32).
Author information
Authors and Affiliations
Contributions
Z.Y. conceived and supervised the project. H.S., T.X. and Z.Y. designed the computational experiments and data analyses. H.S. and X.W. prepared the data. H.S., T.X. and W.G. implemented the methods, conducted the experiments and performed data analyses. H.S., T.X. and Z.Y. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Yi Xiong, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Tables 1–4.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, T., Shi, H., Gao, W. et al. Reusability report: Uncovering associations in biomedical bipartite networks via a bilinear attention network with domain adaptation. Nat Mach Intell 6, 461–466 (2024). https://doi.org/10.1038/s42256-024-00822-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-024-00822-w