Abstract
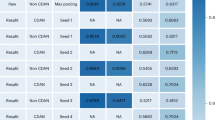
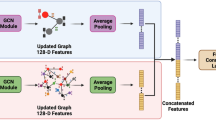
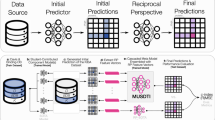
Predicting drug–target interaction is key for drug discovery. Recent deep learning-based methods show promising performance, but two challenges remain: how to explicitly model and learn local interactions between drugs and targets for better prediction and interpretation and how to optimize generalization performance of predictions on novel drug–target pairs. Here, we present DrugBAN, a deep bilinear attention network (BAN) framework with domain adaptation to explicitly learn pairwise local interactions between drugs and targets, and adapt in response to out-of-distribution data. DrugBAN works on drug molecular graphs and target protein sequences to perform prediction, with conditional domain adversarial learning to align learned interaction representations across different distributions for better generalization on novel drug–target pairs. Experiments on three benchmark datasets under both in-domain and cross-domain settings show that DrugBAN achieves the best overall performance against five state-of-the-art baseline models. Moreover, visualizing the learned bilinear attention map provides interpretable insights from prediction results.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The experimental data used in this work are available at https://github.com/peizhenbai/DrugBAN/tree/main/datasets. All data used in this work are from public resources. The BindingDB48 source can be found at https://www.bindingdb.org/bind/index.jsp; the BioSNAP17,30 source can be found at https://github.com/kexinhuang12345/MolTrans/tree/master/dataset/BIOSNAP/full_data and the Human31 source used in a previous study16 can be found at https://github.com/lifanchen-simm/transformerCPI/blob/master/Human%2CC.elegans/dataset/human_data.txt. The co-crystalized ligands from PDB40 are available at https://www.rcsb.org by searching their PDB IDs.
Code availability
The source code and implementation details of DrugBAN are freely available at both GitHub repository (https://github.com/peizhenbai/DrugBAN) and CodeOcean capsule (https://doi.org/10.24433/CO.3558316.v1)57. The code is also archived at Zenodo (https://doi.org/10.5281/zenodo.7231657)58.
References
Luo, Y. et al. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8, 1–13 (2017).
Öztürk, H., Olmez, E. O. & Özgür, A. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics 34, i821–i829 (2018).
Yamanishi, Y., Araki, M., Gutteridge, A., Honda, W. & Kanehisa, M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 24, i232 – i240 (2008).
Zitnik, M. et al. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf. Fusion 50, 71–91 (2019).
Bagherian, M. et al. Machine learning approaches and databases for prediction of drug-target interaction: a survey paper. Brief. Bioinform. 22, 247–269 (2021).
Wen, M. et al. Deep-learning-based drug-target interaction prediction. J. Proteome Res. 16, 1401–1409 (2017).
Sieg, J., Flachsenberg, F. & Rarey, M. In need of bias control: evaluating chemical data for machine learning in structure-based virtual screening. J. Chem. Inf. Model. 59, 947–961 (2019).
Lim, S. et al. A review on compound-protein interaction prediction methods: data, format, representation and model. Comput. Struct. Biotechnol. J. 19, 1541–1556 (2021).
Gao, K. Y. et al. Interpretable drug target prediction using deep neural representation. In Int. Joint Conf. on Artificial Intelligence (IJCAI) 3371–3377 (2018).
Bredel, M. & Jacoby, E. Chemogenomics: an emerging strategy for rapid target and drug discovery. Nat. Rev. Genet. 5, 262–275 (2004).
Lee, I., Keum, J. & Nam, H. DeepConv-DTI: prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Comput. Biol. 15, e1007129 (2019).
Hinnerichs, T. & Hoehndorf, R. DTI-Voodoo: machine learning over interaction networks and ontology-based background knowledge predicts drug-target interactions. Bioinformatics 37, 4835–4843 (2021).
Nguyen, T. et al. GraphDTA: predicting drug-target binding affinity with graph neural networks. Bioinformatics 37, 1140–1147 (2021).
Tsubaki, M., Tomii, K. & Sese, J. Compound protein interaction prediction with end to end learning of neural networks for graphs and sequences. Bioinformatics 35, 309–318 (2019).
Feng, Q., Dueva, E., Cherkasov, A. & Ester, M. PADME: a deep learning-based framework for drug-target interaction prediction. Preprint at arXiv https://arxiv.org/abs/1807.09741 (2018).
Chen, L. et al. TransformerCPI: improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36, 4406–4414 (2020).
Huang, K., Xiao, C., Glass, L. & Sun, J. MolTrans: molecular interaction transformer for drug-target interaction prediction. Bioinformatics 37, 830–836 (2021).
Schenone, M., Dancík, V., Wagner, B. K. & Clemons, P. A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 9, 232–40 (2013).
Öztürk, H., Ozkirimli, E. & Özgür, A. WideDTA: prediction of drug-target binding affinity. Preprint at arXiv https://arxiv.org/abs/1902.04166 (2019).
Zheng, S., Li, Y., Chen, S., Xu, J. & Yang, Y. Predicting drug-protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2, 134–140 (2020).
Abbasi, K. et al. DeepCDA: deep cross-domain compound–protein affinity prediction through lstm and convolutional neural networks. Bioinformatics 36, 4633–4642 (2020).
Kao, P.-Y., Kao, S.-M., Huang, N.-L. & Lin, Y.-C. Toward drug-target interaction prediction via ensemble modeling and transfer learning. In IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM) 2384–2391 (2021).
Abbasi, K., Razzaghi, P., Poso, A., Ghanbari-Ara, S. & Masoudi-Nejad, A. Deep learning in drug target interaction prediction: current and future perspectives. Curr. Med. Chem. 28, 2100–2113 (2021).
Kipf, T. & Welling, M. Semi-supervised classification with graph convolutional networks. In Int. Conf. on Learning Representations (ICLR, 2017).
Yu, Z., Yu, J., Xiang, C., Fan, J. & Tao, D. Beyond bilinear: generalized multimodal factorized high-order pooling for visual question answering. IEEE Trans. Neural Netw. Learn. Syst. 29, 5947–5959 (2018).
Kim, J. -H., Jun, J. & Zhang, B. -T. Bilinear attention networks. In Advances in Neural Information Processing Systems (NeurIPS, 2018).
Long, M., Cao, Z., Wang, J. & Jordan, M. I. Conditional adversarial domain adaptation. In Advances in Neural Information Processing Systems (NeurIPS, 2018).
Weininger, D. SMILES, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Liu, T., Lin, Y., Wen, X., Jorissen, R. N. & Gilson, M. K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 35, D198–D201 (2007).
Zitnik, M., Sosič, R., Maheshwari, S. & Leskovec, J. BioSNAP datasets: Stanford biomedical network dataset collection. https://snap.stanford.edu/biodata (2018).
Liu, H., Sun, J., Guan, J., Zheng, J. & Zhou, S. Improving compound-protein interaction prediction by building up highly credible negative samples. Bioinformatics 31, i221–i229 (2015).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Cao, D., Xu, Q. & Liang, Y. Propy: a tool to generate various modes of chou’s pseaac. Bioinformatics 29, 960–962 (2013).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Ho, T. K. Random decision forests. In Int. Conf. on Document Analysis and Recognition, vol. 1, 278–282 (1995).
Ganin, Y. et al. Domain-adversarial training of neural networks. In J. Mach. Learn. Res. 17, 1–35 (2016).
Kazokaitė, J. et al. Engineered carbonic anhydrase vi-mimic enzyme switched the structure and affinities of inhibitors. Sci. Rep. 9, 1–17 (2019).
Rai, G. et al. Discovery and optimization of potent, cell-active pyrazole-based inhibitors of lactate dehydrogenase (ldh). J. Med. Chem. 60, 9184–9204 (2017).
Fenalti, G. et al. Molecular control of δ-opioid receptor signalling. Nature 506, 191–196 (2014).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Jumper, J. M. et al. Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
Pan, S. J. & Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010).
Gong, B., Grauman, K. & Sha, F. Connecting the dots with landmarks: discriminatively learning domain-invariant features for unsupervised domain adaptation. In Int. Conf. on Machine Learning (ICML) 222-230 (2013).
Huang, J., Smola, A., Gretton, A., Borgwardt, K. M. & Schölkopf, B. Correcting sample selection bias by unlabeled data. In Advances in Neural Information Processing Systems (NIPS) 601–608 (2006).
Li, M. et al. DGL-LifeSci: an open-source toolkit for deep learning on graphs in life science. ACS Omega 6, 27233–27238 (2021).
Song, L., Huang, J., Smola, A. & Fukumizu, K. Hilbert space embeddings of conditional distributions with applications to dynamical systems. In Int. Conf. on Machine Learning (ICML) 961–968 (2009).
Song, L. & Dai, B. Robust low rank kernel embeddings of multivariate distributions. In Advances in Neural Information Processing Systems (NIPS) 3228–3236 (2013).
Gilson, M. K. et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–D1053 (2016).
Bai, P. et al. Hierarchical clustering split for low-bias evaluation of drug-target interaction prediction. In IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM) 641–644 (2021).
Wishart, D. S. et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906 (2008).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems (NeurIPS, 2019).
Wang, M. et al. Deep graph library: a graph-centric, highly-performant package for graph neural networks. Preprint at arXiv https://arxiv.org/abs/1909.01315 (2019).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Harris, C. R. et al. Array programming with numpy. Nature 585, 357–362 (2020).
The pandas development team. pandas-dev/pandas: Pandas 1.2.4. Zenodo https://doi.org/10.5281/zenodo.4681666 (2021).
Landrum, G. et al. RDKit: open-source cheminformatics. https://github.com/rdkit/rdkit (2006).
Bai, P., Miljković, F., John, B. & Lu, H. Interpretable bilinear attention network with domain adaptation improves drug-target prediction. CodeOcean https://doi.org/10.24433/CO.3558316.v1 (2022).
Bai, P., Miljković, F., John, B. & Lu, H. peizhenbai/drugban: v1.2.0. Zenodo https://doi.org/10.5281/zenodo.7231657 (2022).
Kim, J.-H. et al. Hadamard product for low-rank milinear pooling. In Int. Conf. on Learning Representations (ICLR, 2017).
Acknowledgements
We thank S. Zhou, X. Liu and L. Schöbs for their helpful suggestions and discussion on the work. P.B. is supported by a University of Sheffield Faculty of Engineering Research Scholarship (grant no. 169426530).
Author information
Authors and Affiliations
Contributions
P.B., F.M., B.J. and H.L. conceived and designed the work. P.B. developed the models and performed the experiments under the guidance of B.J. and H.L. F.M. and P.B. analysed the data and conducted method comparisons. F.M. contributed to materials and the analysis tool. All authors contributed to writing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Background Information; Discussion; Figs. 1–3; Tables 1–7.
Source data
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bai, P., Miljković, F., John, B. et al. Interpretable bilinear attention network with domain adaptation improves drug–target prediction. Nat Mach Intell 5, 126–136 (2023). https://doi.org/10.1038/s42256-022-00605-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00605-1
This article is cited by
-
CAT-DTI: cross-attention and Transformer network with domain adaptation for drug-target interaction prediction
BMC Bioinformatics (2024)
-
Reusability report: Uncovering associations in biomedical bipartite networks via a bilinear attention network with domain adaptation
Nature Machine Intelligence (2024)
-
Predicting emerging drug interactions using GNNs
Nature Computational Science (2023)
-
ZeroBind: a protein-specific zero-shot predictor with subgraph matching for drug-target interactions
Nature Communications (2023)
-
Classification models for predicting the bioactivity of pan-TRK inhibitors and SAR analysis
Molecular Diversity (2023)