Abstract
High throughput screening (HTS) is routinely used to identify bioactive small molecules. This requires physical compounds, which limits coverage of accessible chemical space. Computational approaches combined with vast on-demand chemical libraries can access far greater chemical space, provided that the predictive accuracy is sufficient to identify useful molecules. Through the largest and most diverse virtual HTS campaign reported to date, comprising 318 individual projects, we demonstrate that our AtomNet® convolutional neural network successfully finds novel hits across every major therapeutic area and protein class. We address historical limitations of computational screening by demonstrating success for target proteins without known binders, high-quality X-ray crystal structures, or manual cherry-picking of compounds. We show that the molecules selected by the AtomNet® model are novel drug-like scaffolds rather than minor modifications to known bioactive compounds. Our empirical results suggest that computational methods can substantially replace HTS as the first step of small-molecule drug discovery.
Similar content being viewed by others
Introduction
Despite present interest in AI/ML and thirty years of case studies1,2,3,4, computational screening techniques have achieved limited adoption within the pharmaceutical industry. A recent investigation into the origins of 156 clinical candidates5 found that only 1% came from virtual screening; in contrast, over 90% of clinical candidates were derived from patent busting or high throughput screening (HTS). Unfortunately, these sources are increasingly challenged, given the pharmaceutical industry’s shift to novel target classes, such as proximity-induced protein degradation6, protein–protein interactions7, and RNA targeting8.
Currently, HTS is the critical tool in drug discovery, providing most novel scaffolds of recent clinical candidates5,9,10. These initial starting points crucially shape the course of downstream medicinal chemistry efforts, as most drugs preserve at least 80% of the scaffold of the initially identified lead11. Despite these foundational contributions, HTS suffers from practical limitations. Principally, HTS, like all physical experiments, requires that the compounds exist. However, with the advent of synthesis-on-demand libraries, most commercially-available molecules have yet to be synthesized. Still, they can be made and delivered for testing in a matter of weeks12,13,14. These libraries comprise trillions of molecules14,15 that exemplify millions of otherwise-unavailable scaffolds12, providing an opportunity to substantially expand the scope and diversity of available chemical space explored in the standard drug discovery process.
Computational approaches unlock this opportunity by reversing the requirement to make molecules before testing them. When computational experiments replace HTS as the primary screen, molecules are tested before they are made, and the results from these experiments can inform which molecules are worth synthesizing. Computational experiments further promise to improve upon HTS in terms of cost, speed, need to produce significant quantities of protein16, effort of miniaturizing assay formats while maintaining experimental integrity17,18,19, and reducing false-positive and false-negative rates16,20,21,22,23 including artifacts from aggregation, covalent modification of the target, autofluorescence, or interactions with the reporter rather than the target20,24,25. Historical computational techniques such as ligand-based QSAR26,27,28, structure-based docking29,30, and machine learning31,32 purport to address these limitations of physical screening methods. Unfortunately, these techniques have not replaced HTS; in fact, despite increasing interest in ML, the proportion of drugs discovered with computational techniques has remained steady over the past decades5,10.
Because there will always be individual targets for which one screening technique can identify more hits than another, the key question governing if computation is ready to be the default hit discovery technique is whether computational screens can identify hits successfully across a broad range of diverse targets. Unfortunately, despite excellent benchmark accuracies33,34,35, prospective discovery accuracy remains modest33,36,37. For example, Cerón-Carrasco38 reported over 700 virtual screens against the SARS-CoV-2 main protease. However, when the author sought to validate the computational predictions via physical experiments, the identified compounds were barely active (800uM). Computational approaches have also been limited by a need for extensive target-specific training data31,39,40,41, a requirement for high-quality X-ray crystal structures42,43, dependence on human adjudication (so-called ‘cherry-picking’)12, or a limited domain of applicability44,45,46,47,48. Even recent systems have demonstrated utility only in identifying minor variants of known molecules for well-studied proteins with tens of thousands of known binders in their training data49,50. Figure 1 exemplifies the striking similarities between recently ML-developed compounds and their preceding published chemical matter. This is particularly concerning, as a myopic focus on well-studied proteins has been identified as a cause of low productivity in pharmaceutical discovery51.
Pairs of representative compounds extracted from AI patents (right) and corresponding prior patents (left) for clinical-stage programs (CDK792,93, A2Ar-antagonist94,95, MALT196,97, QPCTL98,99, USP1100,101, and 3CLpro102,103). The identical atoms between the chemical structures are highlighted in red.
Nevertheless, we have observed that deep learning approaches are not as limited as these historical examples would imply. Using our AtomNet52,53,54 screening system, we have previously reported success in finding novel scaffolds for targets without known ligands55,56,57, X-ray crystal structures56,57,58,59,60, or both56,57, as well as challenging modulation via protein–protein interaction59,61 or allosteric binding60 (see Supplementary Table S1 for examples). However, individual examples do not demonstrate the overall success of such deep learning systems. We therefore report our internal discovery efforts against 22 targets of pharmaceutical interest. We then attempted to further assess the generalizability and robustness of deep learning predictive systems by identifying bioactive molecules for a diverse set of targets. We partnered with 482 academic labs and screening centers, from 257 different academic institutions across 30 countries, through our academic collaboration program, the Artificial Intelligence Molecular Screen (AIMS). This collaboration afforded an opportunity to prospectively evaluate the utility of the AtomNet model as a primary screen across a broad range of diverse, challenging, and realistic targets. In aggregate, we report successes and failures from 318 prospective experiments and evaluate our AtomNet machine-learning technology’s ability to serve as a viable alternative to physical HTS campaigns.
Results
We investigated the ability of deep learning-based methods to identify novel bioactive chemotypes by applying the AtomNet model to identify hits for 22 internal targets of pharmaceutical interest. We also explored the breadth of applicability of this approach by attempting to identify drug-like hits in single-dose screens for 296 academic targets, of which 49 were followed up with dose–response experiments, and 21 were further validated by exploring analogs of the initial hits. The average hit rate for our internal projects (6.7%) was comparable to the hit rate for our academic collaborations (7.6%).
Internal portfolio validation
As part of Atomwise’s internal drug discovery efforts, we used the AtomNet model instead of high-throughput or DNA-encoded library (DEL) screening. We screened a 16-billion synthesis-on-demand chemical space62, which is several thousand times larger than HTS libraries and even exceeds the size of most DELs without suffering limitations of DNA-compatible chemistry16,23. Each screen requires over 40,000 CPUs, 3,500 GPUs, 150 TB of main memory, and 55 TB of data transfers. We describe the protocol in detail in the Methods section; briefly, we computationally scored each catalog compound after removing molecules that were prone to interfere with the assays or were too similar to known binders of the target or its homologs. The neural network analyzes and scores the 3D coordinates of each generated protein–ligand co-complex, producing a list of ligands ranked by their predicted binding probability. Our workflow then clusters the top-ranked molecules to ensure diversity and algorithmically selects the highest-scoring exemplars from each cluster. At no point are compounds manually cherry-picked. The molecules were synthesized at Enamine (https://enamine.net) and quality controlled by LC–MS to purity > 90%, in agreement with HTS standards63. Hits were further validated using NMR. We then physically tested, on average, 440 compounds per target at reputable contract research organizations (CROs), while attempting to mitigate assay interferences such as aggregation and oxidation with standard additives (e.g., Tween-20, Triton-X 100, and dithiothreitol (DTT)). We describe the assay protocols in detail in the Supplementary Data S1.
We describe the results of the 22 experiments in Table 1. In 91% of the experiments, we identified single-dose (SD) hits that were reconfirmed in dose–response (DR) experiments. The average target DR hit rate was 6.7% compared to 8.8% from the SD screens. Only 16 of the 22 projects were structurally enabled with X-ray crystallography; one used a cryo-EM structure, while five used homology models with an average sequence identity of 42% to their template protein. The DR hit rate for the cryo-EM project was 10.56%, while the average hit rate for the homology models was a similar 10.8%.
We then advanced 14 projects with at least one dose-responsive scaffold to a round of analog expansion. We found new bioactive analogs in the SD screen for all projects, with an average hit rate of 29.8%. Further validation with DR resulted in an average hit rate of 26% per project, which compares favorably with typical HTS hit rates ranging from 0.151 to 0.001%64,65. We note that the size and chemical diversity within and between physical66 and virtual14 HTS libraries prevent an explicit evaluation of the methods over the same chemical space. The most potent analogs ranged from single-digit nanomolar, against a kinase, to double-digit micromolar, against a transcription factor (Supplementary Table S2). Additionally, we present two internal studies in detail. For Large Tumor Suppressor Kinase 1 (LATS1), we identified potent compounds despite the lack of a crystal structure or known active compounds. For ATP-driven chaperone Valosin Containing Protein (VCP) we identified novel allosteric and orthosteric modulators.
Academic validation
In addition to our internal discovery efforts, we performed virtual screens for 296 targets, comprising more than 20 billion individual neural network scores of generated protein–ligand co-complexes. We purchased, on average, 85 off-the-shelf commercially available compounds, quality controlled by NMR and LC–MS to > 90% purity63, and plated in a single 96-well plate. The compounds were then physically screened for activity against the target of interest in single-dose assays (see Supplemental Data S1 for assay protocols). As with HTS primary screens, additional characterization studies are required to validate the initially identified hits so, in 49 projects, we performed dose–response studies and analog expansion. We present a summary of our results in Supplementary Table S3.
Figure 2 illustrates the distributions of projects across therapeutic areas, protein families, and assay types. Every major therapeutic area is represented, with the most frequent area being oncology, comprising 35% of projects, followed by infectious diseases and neurology, comprising 27% and 9% of projects, respectively. Breaking down the projects by protein families reveals that all major enzyme classes are represented, with enzymes comprising 59% of the targets and membrane proteins such as GPCR, transporters, and ion channels, representing 12% of the targets. Working on a large and diverse set of therapeutic targets requires a heterogeneous collection of biological assays; 20% of the assays measured direct binding, whereas 56% and 20% were functional and phenotypic.
In 215 projects, we identified at least one bioactive compound for the target in a biochemical or cell-based assay. This 73% success rate substantially improves over the ∼50% success rate for HTS21,67. On average, we screened 85 compounds per project and discovered 4.6 active hits, with an average hit rate of 5.5%. For the subset of targets where we found any hits, the average was 6.4 hits per project. Thus, we achieved an average hit rate of 7.6%, which again compares favorably with typical HTS hit rates. See Supplementary Material S1 for all assay definitions and conditions. Supplementary Table S4 shows a representative bioactive compound from each of the 215 successful projects, and Supplementary Fig. S2 shows that the physicochemical properties of the identified hits are largely druglike and Lipinski-compliant.
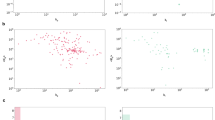
The AtomNet technology robustly identified active molecules, even for targets that lacked prior on-target bioactivity data. This ability to identify hits for previously undrugged targets is critical if machine learning-based approaches are to replace HTS as the default primary screening approach. For 207 out of the 296 targets (70%), the training data available for AtomNet models lacked a single active molecule for that target or any closely related protein (i.e., proteins with sequence identity greater than 70%). We interpret this as evidence of the ability of properly-architected machine learning systems to extrapolate to novel biological space. Figure 3A illustrates the hit rate versus the number of training examples available to our model. Although previous computational approaches typically require thousands of on-target training examples31,39,42, the lack of correlation between training examples and hit rate (R2 = 0.0021, p-value = 0.43) shows that our ML algorithm is agnostic to the availability of such data. We achieved an average success rate of 75% and hit rates of 5.3% when no training data was available, comparable to the 67% and 6.1% success and hit rates achieved when binding data was available in the training set. Interestingly, we also do not see a significant increase in hit rate attributable to the proportion of binding data available for a target (R2 = 0.008, p-value = 0.39). This reflects the robustness of the screening protocol and the chemical dissimilarity of scaffolds identified by AtomNet models to previously known bioactive compounds.
(A) An illustration of the hit rate versus the number of training examples available to our model. Each point represents a project, with the x-axis denoting the number of active molecules in our training for the target protein or homologs and the y-axis denoting the hit rate of the project (the percentage of molecules tested in the project that were active). The model shows no dependence on the availability of on-target training examples. For 70% of the targets, the AtomNet model training data lacked any active molecules for that target or any similar targets with greater than 70% sequence identity, yet the model achieved a hit rate of 5.3% compared to 6.1% when on-target data was available. (B) The distribution of similarities between hits and their most-similar bioactive compounds in our training data. Our screening protocol ensures that the compounds subjected to physical testing are not similar to known active compounds or close homologs (< 0.5 Tanimoto similarity using ECFP4, 1024 bits). Because 70% of the AIMS targets had no annotated bioactivities in our training dataset, hits identified in these projects have a similarity value of zero.
Next, we assessed the ability of the AtomNet models to identify novel scaffolds. This is a critical capability for primary screens, as follow-up assays tend to work within the chemical space uncovered in the initial screen. The task of novel scaffold identification appears in two distinct scenarios: (1) when no scaffold is known for the target and we wish to identify the first scaffold, and (2) when some scaffolds are known but we wish to identify dissimilar scaffolds because novel chemical matter can yield improved selectivity, toxicity, pharmacokinetics, or patentability. Performance of AtomNet models for the first scenario, when no scaffolds for the target existed in the AtomNet model training data, was evaluated on 70% of the targets, where the training data contained no active molecules for the target or its homologs (vide supra). We achieved an average hit rate of 5.3% for targets with no training data. For the second scenario, we analyzed the similarity of the identified hits to known bioactive compounds in our training data (Fig. 3B). Our screening protocol ensures that the compounds subjected to physical testing are not similar to known active compounds or close homologs (< 0.5 Tanimoto similarity using ECFP468, 1024 bits). We interpret this as evidence of the ability of properly-architected machine learning systems to extrapolate to novel chemical space as well. For cases where training data was available (i.e., the Tanimoto similarity is above zero), the similarity distribution is close to the one expected by random compound pairs69. The novelty of the small-molecule structures is striking because target-specific machine-learning algorithms tend to uncover highly similar analogs for known bioactive molecules50,70,71. The superior performance of the AtomNet model is expected, considering the bias-variance tradeoff72 in machine learning algorithms. Because the AtomNet convolutional neural network is a global model, concurrently trained on millions of bioactivities, hundreds of thousands of small molecules, and thousands of protein binding sites, it can reduce both bias and variance of the model compared to target-specific ones33. Specifically, our global model can benefit from multiple levels of information captured in the structures of the small molecules, the sequences of the target proteins, and the three-dimensional interactions between the two.
AtomNet also successfully identified active molecules when there was no X-ray crystal structure of the receptor. Figure 4A compares the hit rates obtained with 3-dimensional crystal structures, cryo-EM, and homology modeling. We did not attempt to select targets based on the similarity to the template but rather used the best template available. We observe no substantial difference in success rate between the three, in contrast to the common challenges in using homology models or low-precision structures for structure-based discovery42,43,73. We achieved average hit rates of 5.6%, 5.5%, and 5.1% for crystal structures, cryo-EM, and homology modeling. We also successfully identified active compounds in projects with NMR structures, but the number of such targets is too small to make statistically-robust claims.
Hit rates obtained for the 296 AIMS projects. (A) A comparison of hit rates using X-ray crystallography, NMR, Cryo-EM, and homology for modeling the structure of the proteins. Each point represents a project with the x-axis denoting the hit rate of the project (the percentage of molecules tested in the project that were active). The number of projects of each type is given in parentheses. We observed no substantial difference in success rate between the physical and the computationally inferred models. We achieved average hit rates of 5.6%, 5.5%, and 5.1% for crystal structures, cryo-EM, and homology modeling, respectively. The number of projects using NMR structures is too small to make statistically-robust claims. (B) A comparison of hit rates observed for traditionally challenging target classes such as protein–protein interactions (PPI) and allosteric binding. Of the 296 projects, 72 targeted PPIs and 58 allosteric binding sites. The average hit rates were 6.4% and 5.8% for PPIs and allosteric binding, respectively. (C) Comparison of hit rates observed for different target classes and (D) enzyme classes. No protein or enzyme class falls outside the domain of applicability of the algorithm.
An interesting demonstration of the robustness of the AtomNet model to low data and poorly characterized protein structure is its ability to identify novel hits for traditionally challenging target classes such as protein–protein interaction (PPI) sites and allosteric binding sites (Fig. 3B). Of the 296 projects, 72 targeted PPIs and 58 allosteric binding sites. We identified hits for 53 (74%) PPI sites and 46 (79%) allosteric sites, with 13 projects representing allosteric sites at PPI interfaces. The average hit rate was 6.4% and 5.8% for PPIs and allosteric binding sites, respectively. The algorithm's success in these target classes, which often suffer from poorly characterized binding sites and a lack of bioactivity training data, is not surprising because Fig. 2A shows that our model is largely not dependent on the availability of on-target training data.
Finally, we investigated whether the algorithm exhibits domain of applicability limitations regarding different protein classes. Figures 4C and 3D illustrate the hit rate observed for each protein and enzyme class. No protein or enzyme class falls outside the domain of applicability of the algorithm, demonstrating that machine learning-based approaches are well-suited as a default technology for new scaffold identification. The hit rate for nuclear receptors is an outlier, with seemingly better accuracy than other classes, but a single data point is not statistically meaningful.
Dose–response validation studies
We performed additional validation studies for 49 AIMS projects with at least one reported hit. The objective of the validation studies was to establish dose–response (DR) relationships for the single-dose (SD) hits. We describe the protocol of the DR experiments in the Methods section. Briefly, we performed dose–response measurements for the reported hits from the single-dose primary screens. DR was determined using the same assay and screening protocol as the single-dose screens, at the same lab, and with the same personnel. Full dose response curves were obtained in most cases, however in some instances a full curve was not obtained, or concentration dependent activity was qualitatively determined by testing at concentrations other than that for the primary screen. The distribution of assay types and target classes for the projects selected for DR validation also was similar to that of the AIMS projects (Supplementary Fig. S3).
We describe the results of the DR experiments in Supplementary Table S5. In 84% of the experiments, we validated at least one SD hit and got a DR readout. The median activity for the total of 144 DR measurements was 15.4 µM (which compares favorably with HTS25,74), of which 13% showed sub-µM potency. Overall, we achieved an average of 2.8 hits per validation study, resulting in a hit rate of 51%. The false positive rate of 49% observed in these experiments is favorably compared to HTS’ which can be as high as 95%20,75. This difference in false positive rates may stem from the comparative ease and robustness of the low-throughput assay format we employed versus high-throughput assay. Representative dose–response curves for each of the 49 projects are shown in Supplementary Table S6.
Analog validation studies
For a subset of 21 projects, we further validated hits with DR activity by testing analogs of the active compounds. In those cases, we used the AtomNet platform to search a purchasable space for additional bioactive compounds chemically analogous to the SD hits. We selected up to 35 additional compounds for testing, including the active compounds from the SD screens.
We describe the results of the analoging experiments in Supplementary Table S7. We identified additional analogs with DR readouts for 16 projects (76%). The median DR activity of the 154 validated analogs was 7.4 µM compared to the median of 15.4 µM of the parent compound (Supplementary Fig. S4).
Methods
Screening protocols
AIMS screening protocol
We began by evaluating screening libraries of millions of catalog compounds from commercial vendors MCule (10 M)76 and Enamine in-stock (2.5 M)77. We then selected a drug-like subset via algorithmic filtering by applying Eli Lilly medicinal chemistry filters78 and removing likely false positives, such as aggregators, autofluorescers, and PAINS79 (see Fig. 2 for the distributions of drug-like properties of the SD hits). The resulting library was virtually screened against the target of interest, removing any molecules with greater than 0.5 Tanimoto similarity in ECFP4 space to any known binders of the target and its homologs within 70% sequence identity. For kinase targets, we extend the exclusion to the whole kinome. The binding site was defined using co-complexes, mutagenesis studies, co-complexes of homologs, or by identifying potential sites using ICM Pocket Finder80 or Fpocket81. Some were orthosteric, while others were allosteric, or as yet unestablished biological functions. In 64 cases, we built homology models using the closest sequence, with an average sequence similarity of 54%. We clustered the top 30,000 molecules using the Butina82 algorithm with a Tanimoto similarity cutoff of 0.35 in ECFP4 space, selecting the highest-scoring exemplars. Additional computed physico-chemical property filters were applied as needed. At no point were compounds cherry-picked. We purchased, on average, 85 compounds, quality controlled by LC–MS to > 90% purity, generally dispensed as 10 mM DMSO stocks plated in a single 96-well plate. In addition, two vials of DMSO-only negative controls were included before scrambling the compound locations on the plate, by the supplier, for blinded experimental testing. To further control for potential artifacts, we removed compounds that showed measurable activity toward more than one target from the analysis.
Dose–response and analoging validation screening protocol
We considered advancing AIMS projects to additional validation studies based on the ability to reorder at least some of the initial SD hits, the availability of chemical analogs in the screening library to the initial hits, the capability to perform dose–response experiments, and the ability of the collaborators to perform additional screens and return results promptly.
We performed two sets of experiments: DR validation of the SD hits from AIMS and analoging with DR readouts. We performed DR measurements using the same assays and protocols as SD.
We performed an analoging round by identifying, for each AIMS hit, its 1000 nearest neighbors from the Mcule library76, using molecular fingerprints similarity68. We augmented the set with additional analogs using substructure83 or FTrees84 searches, if needed. We used an AtomNet regression model, trained to predict quantitative bioactivities (e.g., IC50 or Ki), to score and rank the analogs. A set of 20—35 compounds from the analogs space of an initial hit were then obtained based on similarity and top scores from the AtomNet model for testing.
Internal portfolio screening protocol
We followed a protocol similar to the AIMS screen with a few deviations. First, we used the Enamine REAL library of over 16 billion compounds62. Second, we used an ensemble of six AtomNet models for the screens. Last, on average, we selected a set of 440 compounds for testing.
The analoging protocol is similar to the AIMS validation studies, with the following deviations. First, we used the Enamine REAL library for analog search. Second, we selected an average of 676 analogs per project. Third, the analog search protocol was more complex, pulling nearest neighbors based on maximum common substructure and graph edit distance in addition to the ECFP4-based one.
AtomNet® model architecture
We previously published in detail52,53,55,58,59,61,85,86 during the course of the AIMS program, and we described the most recent version of the AtomNet model architecture in detail elsewhere53. We provide a brief description below.
The AtomNet model is a Graph Convolution Network architecture with atoms represented as vertices and pair-wise, distance-dependent, edges representing atom proximities. The input is a graph network of features characterizing the atom types and topologies of an ensemble of protein–ligand complexes. Receptor atoms more than 7 Å away from any ligand atom are excluded from the complexes, and each node in the graph is associated with a feature vector representing the atom type using Sybyl typing87.
The network has five graph convolutional blocks. In the first two graph convolution blocks, all ligand and receptor atoms 5 Å apart from each other are considered, and 64 filters per block are used. In the third block, the cutoff radius and filters are increased to 7 Å and 128, respectively. Only ligand features in the last two blocks are considered without changing the threshold cutoff or the number of filters. Finally, the sum-pool of the ligand-only layer creates a 3-task layer on top of the network. That multi-task layer predicts three endpoints: bioactivity, pose quality, and a physics-based docking score88.
We trained an ensemble of 6 models, splitting the training data into sixfold cross-validation sets based on a protein sequence similarity cutoff of 70%. Then, each model in the ensemble was trained on a different fold for 10 epochs, using the ADAM optimizer89 with a learning rate of 0.001, and targets were sampled with replacement, proportional to the number of active compounds associated with that target.
Data
All data generated or analyzed during this study are included in this published article (and its supplementary information S1 files). Boxplots illustrations show the quartiles (Q1 and Q3) of the dataset while the whiskers extend to show the rest of the distribution, except for points that are determined to be “outliers” (1.5 × of the inter-quartile range, as implemented in the Seaborn and Matplotlib toolboxes90,91).
Conclusion
HTS is the most widely-used tool for hit discovery for new targets. Unfortunately, all physical screening methods share the critical limitation that a molecule must exist to be screened. Computational methods enable a fundamental shift to a test-then-make paradigm. In this work, we report on 318 projects (22 internal projects and 296 collaborations) where we used the AtomNet platform as the primary screening tool coupled with low-throughput physical screens as validation. The AtomNet technology can identify bioactive scaffolds across a wide range of proteins, even without known binders, X-ray structures, or manual cherry-picking of compounds. Our empirical results suggest that machine learning approaches have reached a computational accuracy that can replace HTS as the first step of small-molecule drug discovery.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Kuntz, I. D. Structure-based strategies for drug design and discovery. Science 257, 1078–1082 (1992).
Bajorath, J. Integration of virtual and high-throughput screening. Nat. Rev. Drug Discov. 1, 882–894 (2002).
Walters, W. P., Stahl, M. T. & Murcko, M. A. Virtual screening—an overview. Drug Discov. Today 3, 160–178 (1998).
Ring, C. S. et al. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc. Natl. Acad. Sci. USA. 90, 3583–3587 (1993).
Brown, D. G. An analysis of successful hit-to-clinical candidate pairs. J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.3c00521 (2023).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Lu, H. et al. Recent advances in the development of protein–protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 5, 1–23 (2020).
Childs-Disney, J. L. et al. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21, 736–762 (2022).
Brown, D. G. & Boström, J. Where do recent small molecule clinical development candidates come from?. J. Med. Chem. 61, 9442–9468 (2018).
Dragovich, P. S., Haap, W., Mulvihill, M. M., Plancher, J.-M. & Stepan, A. F. Small-molecule lead-finding trends across the roche and genentech research organizations. J. Med. Chem. 65, 3606–3615 (2022).
Perola, E. An analysis of the binding efficiencies of drugs and their leads in successful drug discovery programs. J. Med. Chem. 53, 2986–2997 (2010).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224 (2019).
Sadybekov, A. A. et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature 601, 452–459 (2022).
Bellmann, L., Penner, P., Gastreich, M. & Rarey, M. Comparison of combinatorial fragment spaces and its application to ultralarge make-on-demand compound catalogs. J. Chem. Inf. Model. 62, 553–566 (2022).
Neumann, A., Marrison, L. & Klein, R. Relevance of the trillion-sized chemical space “explore” as a source for drug discovery. ACS Med. Chem. Lett. 14, 466–472 (2023).
Sunkari, Y. K., Siripuram, V. K., Nguyen, T.-L. & Flajolet, M. High-power screening (HPS) empowered by DNA-encoded libraries. Trends Pharmacol. Sci. 43, 4–15 (2022).
Malo, N., Hanley, J. A., Cerquozzi, S., Pelletier, J. & Nadon, R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175 (2006).
Iversen, P. W., Eastwood, B. J., Sittampalam, G. S. & Cox, K. L. A comparison of assay performance measures in screening assays: Signal window, Z’ factor, and assay variability ratio. J. Biomol. Screen. 11, 247–252 (2006).
Zhang, J.-H., Chung, T. D. Y. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999).
Jadhav, A. et al. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem. 53, 37–51 (2010).
Fox, S. et al. High-throughput screening: Update on practices and success. J. Biomol. Screen. 11, 864–869 (2006).
Owen, S. C., Doak, A. K., Wassam, P., Shoichet, M. S. & Shoichet, B. K. Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chem. Biol. 7, 1429–1435 (2012).
Rössler, S. L., Grob, N. M., Buchwald, S. L. & Pentelute, B. L. Abiotic peptides as carriers of information for the encoding of small-molecule library synthesis. Science 379, 939–945 (2023).
McGovern, S. L., Caselli, E., Grigorieff, N. & Shoichet, B. K. A Common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 45, 1712–1722 (2002).
Feng, B. Y., Shelat, A., Doman, T. N., Guy, R. K. & Shoichet, B. K. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1, 146–148 (2005).
Martin, E. J., Polyakov, V. R., Tian, L. & Perez, R. C. Profile-QSAR 2.0: Kinase virtual screening accuracy comparable to four-concentration IC50s for realistically novel compounds. J. Chem. Inf. Model. 57, 2077–2088 (2017).
Keiser, M. J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009).
Svetnik, V. et al. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 43, 1947–1958 (2003).
Kitchen, D. B., Decornez, H., Furr, J. R. & Bajorath, J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 3, 935–949 (2004).
Shoichet, B. K. Virtual screening of chemical libraries. Nature 432, 862–865 (2004).
Ma, J., Sheridan, R. P., Liaw, A., Dahl, G. E. & Svetnik, V. Deep neural nets as a method for quantitative structure-activity relationships. J. Chem. Inf. Model. 55, 263–274 (2015).
Sheridan, R. P. et al. Machine Learning and Deep Learning Experimental error, kurtosis, activity cliffs, and methodology: What limits the predictivity of QSAR models?. J. Chem. Inf. Model. https://doi.org/10.1021/acs.jcim.9b01067 (2020).
Wallach, I. & Heifets, A. Most ligand-based classification benchmarks reward memorization rather than generalization. J. Chem. Inf. Model. 58, 916–932 (2018).
Chen, L. et al. Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLOS ONE 14, e0220113 (2019).
Chuang, K. V. & Keiser, M. J. Comment on “Predicting reaction performance in C–N cross-coupling using machine learning”. Science 362, eaat8603 (2018).
Gaieb, Z. et al. D3R Grand Challenge 3: Blind prediction of protein–ligand poses and affinity rankings. J. Comput. Aided Mol. Des. 33, 1–18 (2019).
Gabel, J., Desaphy, J. & Rognan, D. Beware of machine learning-based scoring functions on the danger of developing black boxes. J. Chem. Inf. Model. 54, 2807–2815 (2014).
Cerón-Carrasco, J. P. When virtual screening yields inactive drugs: dealing with false theoretical friends. ChemMedChem 17, e202200278 (2022).
McCloskey, K. et al. Machine learning on DNA-encoded libraries: A new paradigm for hit-finding. J. Med. Chem. 63, 8857–8866 (2020).
Wenzel, J., Matter, H. & Schmidt, F. Predictive multitask deep neural network models for ADME-Tox properties: Learning from large data sets. J. Chem. Inf. Model. 59, 1253–1268 (2019).
Feinberg, E. N. et al. PotentialNet for molecular property prediction. ACS Cent. Sci. 4, 1520–1530 (2018).
Schindler, C. E. M. et al. Large-scale assessment of binding free energy calculations in active drug discovery projects. J. Chem. Inf. Model. 60, 5457–5474 (2020).
Bordogna, A., Pandini, A. & Bonati, L. Predicting the accuracy of protein–ligand docking on homology models. J. Comput. Chem. 32, 81–98 (2011).
Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell 180, 688-702.e13 (2020).
Melo, M. C. R., Maasch, J. R. M. A. & de la Fuente-Nunez, C. Accelerating antibiotic discovery through artificial intelligence. Commun. Biol. 4, 1–13 (2021).
Skinnider, M. A. et al. A deep generative model enables automated structure elucidation of novel psychoactive substances. Nat. Mach. Intell. 3, 973–984 (2021).
Muegge, I. & Oloff, S. Advances in virtual screening. Drug Discov. Today Technol. 3, 405–411 (2006).
N. Muratov, E. et al. QSAR without borders. Chem. Soc. Rev. 49, 3525–3564 (2020).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Walters, W. P. & Murcko, M. Assessing the impact of generative AI on medicinal chemistry. Nat. Biotechnol. 38, 143–145 (2020).
Scannell, J. W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191 (2012).
Wallach, I., Dzamba, M. & Heifets, A. AtomNet: A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery. ArXiv Prepr. ArXiv151002855 1–11 (2015).
Gniewek, P., Worley, B., Stafford, K., van den Bedem, H. & Anderson, B. Learning physics confers pose-sensitivity in structure-based virtual screening. https://doi.org/10.48550/arXiv.2110.15459 (2021).
Stafford, K. A., Anderson, B. M., Sorenson, J. & van den Bedem, H. AtomNet PoseRanker: Enriching ligand pose quality for dynamic proteins in virtual high-throughput screens. J. Chem. Inf. Model. 62, 1178–1189 (2022).
Hsieh, C.-H. et al. Miro1 marks parkinson’s disease subset and miro1 reducer rescues neuron loss in Parkinson’s models. Cell Metab. 30, 1131-1140.e7 (2019).
Reidenbach, A. G. et al. Multimodal small-molecule screening for human prion protein binders. J. Biol. Chem. 295, 13516–13531 (2020).
Bon, C. et al. Discovery of novel trace amine-associated receptor 5 (TAAR5) antagonists using a deep convolutional neural network. Int. J. Mol. Sci. 23, 3127 (2022).
Stecula, A., Hussain, M. S. & Viola, R. E. Discovery of novel inhibitors of a critical brain enzyme using a homology model and a deep convolutional neural network. J. Med. Chem. 63, 8867–8875 (2020).
Su, S. et al. SPOP and OTUD7A Control EWS–FLI1 protein stability to govern ewing sarcoma growth. Adv. Sci. 8, 2004846 (2021).
Pedicone, C. et al. Discovery of a novel SHIP1 agonist that promotes degradation of lipid-laden phagocytic cargo by microglia. iScience 25, 104170 (2022).
Huang, C. et al. Small molecules block the interaction between porcine reproductive and respiratory syndrome virus and CD163 receptor and the infection of pig cells. Virol. J. 17, 116 (2020).
Grygorenko, O. O. et al. Generating multibillion chemical space of readily accessible screening compounds. iScience 23, 101681 (2020).
Dandapani, S., Rosse, G., Southall, N., Salvino, J. M. & Thomas, C. J. Selecting, acquiring, and using small molecule libraries for high-throughput screening. Curr. Protoc. Chem. Biol. 4, 177–191 (2012).
Schuffenhauer, A. et al. Library design for fragment based screening. Curr. Top. Med. Chem. 5, 751–762 (2005).
Jacoby, E. et al. Key aspects of the novartis compound collection enhancement project for the compilation of a comprehensive Chemogenomics drug discovery screening collection. Curr. Top. Med. Chem. 5, 397–411 (2005).
Petrova, T., Chuprina, A., Parkesh, R. & Pushechnikov, A. Structural enrichment of HTS compounds from available commercial libraries. MedChemComm 3, 571–579 (2012).
Macarron, R. et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 10, 188–195 (2011).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Riniker, S. & Landrum, G. A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminformatics 5, 26 (2013).
Ren, F. et al. AlphaFold accelerates artificial intelligence powered drug discovery: Efficient discovery of a novel cyclin-dependent kinase 20 (CDK20) Small Molecule Inhibitor (2022).
Assessing structural novelty of the first AI-designed drug candidates to go into human clinical trials. CAS https://www.cas.org/resources/blog/ai-drug-candidates.
Kohavi, R. & Wolpert, D. Bias plus variance decomposition for zero-one loss functions. in Proceedings of the Thirteenth International Conference on International Conference on Machine Learning 275–283 (Morgan Kaufmann Publishers Inc., San Francisco, CA, USA, 1996).
Ferrara, P. & Jacoby, E. Evaluation of the utility of homology models in high throughput docking. J. Mol. Model. 13, 897–905 (2007).
Walters, W. P. & Namchuk, M. Designing screens: How to make your hits a hit. Nat. Rev. Drug Discov. 2, 259–266 (2003).
Inglese, J. et al. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3, 466–479 (2007).
mcule database. https://mcule.com/database/.
Screening Collections - Enamine. https://enamine.net/compound-collections/screening-collection.
Bruns, R. F. & Watson, I. A. Rules for identifying potentially reactive or promiscuous compounds. J. Med. Chem. 55, 9763–9772 (2012).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Abagyan, R. & Kufareva, I. The flexible pocketome engine for structural chemogenomics. Methods Mol. Biol. Clifton NJ 575, 249–279 (2009).
Le Guilloux, V., Schmidtke, P. & Tuffery, P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics 10, 168 (2009).
Butina, D. Unsupervised data base clustering based on daylight’s fingerprint and tanimoto similarity: A fast and automated way to cluster small and large data sets. J. Chem. Inf. Comput. Sci. 39, 747–750 (1999).
RDKit: Open-Source Cheminformatics.
Rarey, M. & Dixon, J. S. Feature trees: A new molecular similarity measure based on tree matching. J. Comput. Aided Mol. Des. 12, 471–490 (1998).
Stafford, K., Anderson, B. M., Sorenson, J. & van den Bedem, H. AtomNet PoseRanker: Enriching Ligand Pose Quality for Dynamic Proteins in Virtual High Throughput Screens. https://doi.org/10.26434/chemrxiv-2021-t6xkj (2021).
Schroedl, S. Current methods and challenges for deep learning in drug discovery. Drug Discov. Today Technol. 32–33, 9–17 (2019).
Bender, A., Mussa, H. Y., Glen, R. C. & Reiling, S. Molecular similarity searching using atom environments, information-based feature selection, and a Naïve Bayesian classifier. J. Chem. Inf. Comput. Sci. 44, 170–178 (2004).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization. ArXiv14126980 Cs (2017).
Waskom, M. L. seaborn: Statistical data visualization. J. Open Source Softw. 6, 3021 (2021).
Hunter, J. D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Marineau, J. J. et al. Discovery of SY-5609: A selective, noncovalent inhibitor of CDK7. J. Med. Chem. 65, 1458–1480 (2022).
Gu, X., BAI, H., Barbeau, O. R. & Besnard, J. Aromatic heterocyclic compound, and pharmaceutical composition and application thereof. (2022).
Barbay, J. K., Chakravarty, D., Leonard, K., Shook, B. C. & Wang, A. Phenyl and heteroaryl substituted thieno[2,3-d]Pyrimidines and their use as adenosine A2a receptor antagonists (2010).
Bell, A. S., Schreyer, A. M. & Versluys, S. Pyrazolopyrimidine compounds as adenosine receptor antagonists (2019).
Soldermann, C. P. et al. Pyrazolo pyrimidine derivatives and their use as MALT1 inhbitors (2019).
Feng, S. et al. Tricyclic compounds useful in the treatment of cancer, autoimmune and inflammatory disorders (2023).
Heiser, U. & Sommer, R. Inhibitors of glutaminyl cyclase (2020).
Cheng, X., Liu, Y., Qin, L., Ren, F. & Wu, J. Beta-lactam derivatives for the treatment of diseases (2023).
Wylie, A. A. et al. Therapeutic combinations comprising ubiquitin-specific-processing protease 1 (usp1) inhibitors and poly (adp-ribose) polymerase (parp) inhibitors (2021).
Wu, J., Qin, L. & Liu, J. Small molecule inhibitors of ubiquitin specific protease 1 (usp1) and uses thereof 2023).
Stille, J. et al. Design, Synthesis and Biological Evaluation of Novel SARS-CoV-2 3CLpro Covalent Inhibitors. https://doi.org/10.26434/chemrxiv.13087742.v1 (2020).
Zavoronkovs, A., Ivanenkov, Y. A. & Zagribelnyy, B. Sars-cov-2 inhibitors having covalent modifications for treating coronavirus infections. (2021).
Acknowledgements
See Supplementary section S1.
Author information
Authors and Affiliations
Consortia
Contributions
All authors have contributed to the publication, being variously involved in technology development, experimental protocol designs, experimental performance, data acquisition, statistical analysis, and manuscript writing.
Ethics declarations
Competing interests
The authors affiliated with Atomwise declare the existence of a financial competing interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
The Atomwise AIMS Program. AI is a viable alternative to high throughput screening: a 318-target study. Sci Rep 14, 7526 (2024). https://doi.org/10.1038/s41598-024-54655-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54655-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.