Abstract
Tardigrades are microscopic ecdysozoans that can withstand extreme environmental conditions. Several tardigrade species undergo reversible morphological transformations and enter into cryptobiosis, which helps them to survive periods of unfavorable environmental conditions. However, the underlying molecular mechanisms of cryptobiosis are mostly unknown. Tubulins are evolutionarily conserved components of the microtubule cytoskeleton that are crucial in many cellular processes. We hypothesize that microtubules are necessary for the morphological changes associated with successful cryptobiosis. The molecular composition of the microtubule cytoskeleton in tardigrades is unknown. Therefore, we analyzed and characterized tardigrade tubulins and identified 79 tardigrade tubulin sequences in eight taxa. We found three α-, seven β-, one γ-, and one ε-tubulin isoform. To verify in silico identified tardigrade tubulins, we also isolated and sequenced nine out of ten predicted Hypsibius exemplaris tubulins. All tardigrade tubulins were localized as expected when overexpressed in mammalian cultured cells: to the microtubules or to the centrosomes. The presence of a functional ε-tubulin, clearly localized to centrioles, is attractive from a phylogenetic point of view. Although the phylogenetically close Nematoda lost their δ- and ε-tubulins, some groups of Arthropoda still possess them. Thus, our data support the current placement of tardigrades into the Panarthropoda clade.
Similar content being viewed by others
Introduction
Tardigrades, also called water bears, are microscopic ecdysozoans belonging to the phylum Tardigrada, divided into three classes: well-defined Heterotardigrada, Eutardigrada, and dubious Mesotardigrada1. Tardigrades are known for their unique ability to withstand extreme environmental conditions, such as desiccation, vacuum, low/high temperatures and radiation. Several tardigrade species can undergo reversible morphological transformations into so-called tuns2. Tuns are compacted tardigrade bodies with legs withdrawn inward into the body cavity3,4,5. In their tun form, tardigrades enter into cryptobiosis, a state with a barely detectable metabolism6. Cryptobiosis helps tardigrades survive unfavorable environmental conditions, such as those listed above6,7,8,9,10,11,12,13. After the return of favorable external conditions, tardigrades undergo a reverse morphological change and enter the active stage of life11,14. However, underlying molecular mechanisms governing cryptobiosis have only begun to emerge15,16,17,18,19.
The phylogenetic position of the phylum Tardigrada is under discussion. Some authors proposed that tardigrades belong to the animal clade Panarthropoda (euarthropods and onychophorans)20,21,22,23, while others suggested that Tardigrada and Nematoda have a closer phylogenetic relationship than Tardigrada and Arthropoda do24. Despite mounting molecular evidence, the phylogenetic position of the phylum Tardigrada has remained uncertain.
The cytoskeleton provides a system for cellular movement and shape changes. It consists of three prominent protein filament families—actin, intermediate filaments and microtubules. Although each cytoskeletal filament type has different mechanical properties, dynamics, and biological roles, the systems are interrelated25.
Interestingly, Panarthropoda generally possess microtubules and actin filaments but lack cytoplasmic intermediate filaments (IFs)26. The presence of an exoskeleton providing mechanical support might cause the loss of cytoplasmic IFs in the arthropod lineage26,27. In tardigrades, the cytoplasmatic role of IFs was substituted by a novel lamin-derived protein Cytotardin28. Cytotardin does not form scaffold-like structures throughout the cell but belt-like filaments28. It was found in the epidermis and tissues exposed to mechanical stress and might help resist extreme conditions28.
Microtubules are hollow cylinders composed of evolutionarily conserved heterodimers of tubulin proteins and are critical for many cellular processes29. Tardigrade microtubules were observed in an electron microscopy study in claw glands30 and also in recent studies on tardigrade nervous system21,31,32,33,34,35,36,37.
We hypothesize that microtubules might play an essential role in tardigrade physiology and are necessary for morphological changes associated with cryptobiosis. The molecular composition of the microtubule cytoskeleton in tardigrades is unknown. Therefore, we decided to start elucidating this outstanding question by analyzing and characterizing tardigrade tubulins.
Results
To study the molecular composition of the tardigrade microtubule cytoskeleton, we identified and analyzed tardigrade tubulin coding sequences (CDS). We used published transcriptomes and genomes—with annotated predicted CDS—of eight tardigrade taxa (Table 1). We generated three putative tardigrade tubulin homologs datasets based on full-length sequences and tubulin domains (UJEP dataset, CRG dataset, CRG tubulin domains dataset; see Methods). Next, we ran regressive T-coffee38 alignments of found tardigrade tubulin protein sequences with a large dataset published by Findeisen et al.39 containing 3524 tubulin or tubulin-like sequences from 504 species (Supplementary Data 1).
Next, we constructed phylogenetic trees based on the alignments to estimate which tubulin isotypes and isoforms were found in tardigrades. We used maximum likelihood (IQ-TREE) and minimum evolution (FastME) methods. All phylogenetic trees were always rooted using Archaea tubulin-like sequences, not bacterial tubulin homologs (e.g., FtsZ), to minimize the likelihood of homoplasy events due to a larger phylogenetic distance. The tardigrade tubulins we found clustered into α-, β-, γ-, and ε-tubulin isotypes (Fig. 1, Supplementary Data 2; for the unification of nomenclature, see Supplementary Table S1). Notably, we adopted simplified tubulin naming from Findeisen et al.39, where numbers substituted Greek letters: α-tubulin is Tub1, β-tubulin is Tub2, γ-tubulin is Tub3, δ-tubulin is Tub4, ε-tubulin is Tub5, and ζ-tubulin is Tub6. Individual tubulin isoforms are further distinguished by additional letters and numbers, such as Tub2D3 and Tub2D4, two different tardigrade β-tubulin isoforms.
Phylogenetic analysis of tubulin homologs found in tardigrades. (a-f) Phylogenetic trees computed from alignments of tardigrade tubulin homologous sequences with Findeisen et al.39 dataset. (a, c, e) Minimum evolution method (FastMe). (b, d, f) Maximum likelihood method (IQ-TREE). (a, b) UJEP dataset—full tubulin sequences, (c, d) CRG dataset—full tubulin sequences, (e–f) CRG dataset—extracted tubulin domains only. All phylogenetic trees are rooted by an archaeal tubulin-like group. The legend describes color coding. See also Supplementary data 1 and Supplementary Data 2.
This analysis allowed us to identify several divergent non-tubulin (tPr-orf74 and tPr-GFGY01000052.1x) and putative non-tardigrade sequences that we removed manually from the final dataset. For example, Paramacrobiotus richtersi (Murray, 1911) sequences tPr-orf110, tPr-orf67 and tPr-orf77 exhibited 87%, 87% and 61% identity, respectively, to tubulins in Nematocida displodere Luallen, Reinke, Tong, Botts, Félix & Troemel, 201640, a natural microsporidian parasite of Caenorhabditis elegans. Notably, the carnivorous Pam. richtersi can feed on C. elegans, suggesting these sequences are likely contaminations. We also removed several truncated sequences that were identical to longer protein sequences from the same taxa present in the dataset (Supplementary Table S1).
The curated final dataset was aligned to Findeisen et al.39 tubulin dataset using the regressive mode of T-Coffee38. This alignment served as a template for the computation of the final phylogenetic tree (Fig. 2a, Supplementary Data 3) using the maximum-likelihood method (IQ-TREE). The tardigrade tubulins clustered into α-, β-, γ-, and ε-tubulin isotypes as before. Moreover, we were able to distinguish individual isoforms inside the tubulin isotype clusters based on detailed manual sequence analyses (Fig. 2b, Supplementary Data 3).
Identification of tardigrade tubulin isoforms. (a, b) Different visualizations of an identical phylogenetic tree computed from an alignment of the curated tardigrade tubulin dataset with Findeisen et al.39 dataset. (a) Maximum likelihood method (IQ-TREE). (b) Maximum likelihood method (IQ-TREE), non-tardigrade tubulins branches collapsed and branch lengths ignored to simplify visualization. Grey circles indicate SH-aLRT test score69. The legend describes color coding. See also Supplementary data 3 and Supplementary Table 1.
In total, we found three α-, seven β-, one γ-, and one ε-tubulin isoforms (Supplementary Table S1). Based on sequence analysis, we suspect that tEs-orf27 might be another example of contamination. Its sequence exhibited 75% identity to Pollicipes pollicipes, a barnacle where Echiniscoides s. sigismundi (M. Schultze, 1865) is often collected41.
We determined three α-tubulin isoforms that we named Tub1A1, Tub1A2, and Tub1C1. In Tub1A1, the C-terminal tyrosine residue is replaced by valine; the Tub1A2 seems to have a relatively similar C-terminus to vertebrate α-tubulins; and the Tub1C1 possesses a C-terminal tyrosine or rarely phenylalanine, but lacks the conserved previous double glutamate (Fig. 3, Supplementary Data 4).
We found seven β-tubulin isoforms with relatively diverse C-termini compared to α-tubulins (Fig. 4, Supplementary Data 4). While the Tub2A isoform from Echiniscus testudo (Doyère, 1840) is distant from other tardigrade β-tubulin isoforms, the others share a relatively high degree of similarity, including the heterotardigrade Tub2D4 isoform from Ech. testudo and from Ecn. s. sigismundi. A conserved hydrophobic amino acid (AA) at the relative position 371 and alanine at the relative position 295 in the Tub2C isoform are interesting. Non-polar AAs at these positions are conserved in many βIII-tubulins in vertebrates and some invertebrates42,43. However, other conserved vertebrate βIII-tubulin AAs are absent in Tub2C (Supplementary Data 4).
The γ- and ε-tubulins were found as single isoforms in analyzed tardigrade genomes and transcriptomes (Supplementary Data 4) with two exceptions. However, the truncated version of the Ech. testudo γ-tubulin (tEt-orf155, 304 AA) translated in silico from an alternative start codon might correspond to the full-length Ech. testudo γ-tubulin (tEt-orf155, 526 AA). Two ε-tubulin sequences from Ecn. s. sigismundi—orf187 and orf188 (Supplementary Data 4)—also seem to be fragments of one ε-tubulin sequence, corresponding to N- and C-terminal parts, respectively. However, this must be proven by direct isolation and sequencing of the CDS from Ecn. s. sigismundi specimens in the future.
To verify the function of found tubulin sequences we isolated the identified tubulin CDS from tardigrades and expressed them in mammalian cells (for the lack of robust molecular tools for direct expression in tardigrades). We isolated nine out of ten annotated tubulins from Hypsibius exemplaris Gąsiorek, Stec, Morek & Michalczyk, 201844 (Supplementary Table S2). The Tub2D5 (Genbank OWA50044.1), which we could not amplify despite trying different primer pairs (Supplementary Table S3), was the exception. Translated protein sequences from isolated tubulin CDS were almost identical to previously annotated versions—except for γ-tubulin—with few single AA changes reflecting likely intra-species diversity (Supplementary Table S2).
Interestingly, the length of the previously annotated γ-tubulin was 912 AA (Genbank OQV14445.1; Supplementary Table S2). In the large dataset that includes thousands of eukaryotic tubulins39 and other tardigrade γ-tubulins, no γ-tubulins of similar length were found. We suspected a probable mistake in assigning exon–intron boundaries and thus analyzed the whole coding sequence in the contig (Genbank MTYJ01000105.1). Sequence analysis using NetGene245,46, which predicts splice sites, and C. elegans settings (C. elegans belongs to Ecdysozoa as tardigrades) revealed that the putative donor splice site at the boundary of the exon 4 was dubious. The NetGene2 predicted an alternative splice site further downstream. We modified the coding sequence by adding the putative intronic sequence following the annotated exon 4 until the newly predicted donor splice site (CTGAGCAAAG). This modification gave a stop codon in frame with the coding sequence. Based on this adjustment, we designed a reverse primer downstream of the stop codon (Supplementary Table S3) and successfully amplified a functional γ-tubulin from the adult Hys. exemplaris cDNA.
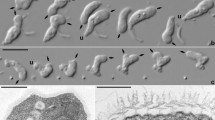
All tardigrade β-tubulin isoforms tagged with mScarlet fluorescent protein localized properly to microtubules (Fig. 5a, b) labelled with human α-tubulin (EGFP-TUBA1B). The mEGFP-tagged tardigrade α-tubulins localized to microtubules as well, as verified by coexpression with tardigrade β-tubulins (Fig. 5c). In general, tardigrade β-tubulins were more easily detected on mammalian microtubules than tardigrade α-tubulins. The weakest microtubule localization was observed with the tHe-Tub1A2. This tardigrade tubulin was mainly cytoplasmic; however, some microtubule localization was detectable (Fig. 5c). The expected subcellular localization was also observed for tardigrade γ- and ε-tubulins, which localized to the centrosomes in human hTert-RPE-1 cells (Fig. 5c). To conclude, all isolated tardigrade tubulins localized to expected subcellular locations.
Tardigrade tubulins localize to microtubules or to the centrosomes in mammalian cultured cells. (a) A schematic of a mammalian cell with microtubules and the centrosome. (b) Maximum intensity projections (MIP) of living U87-MG cells overexpressing indicated constructs. Human EGFP-tagged α-tubulin (TUBA1B) used as a control. Note that all tardigrade β-tubulins localize to microtubules. (c) MIP of living hTert-RPE-1 cells overexpressing indicated constructs. Note that tardigrade γ- and ε-tubulins localize to the centrosome (magenta arrows). Scale bars 10 μm.
Discussion
We found in this study that tardigrades possess four tubulin subfamilies that function similarly to their vertebrate counterparts. Identifying functional ε-tubulin in the eutardigrade Hys. exemplaris and its presence also in heterotardigrades argue in favor of the phylogenetic placement of all tardigrades among Panarthropoda.
Eukaryotic tubulins belong to six major subfamilies (α, β, γ, δ, ε, and ζ), and the last common ancestor of Eukaryota probably had all six tubulin isotypes39,47. We found 23 α-, 43 β-, 9 γ-, and 3 ε-tubulin sequences in tardigrades. The number of β-tubulins is generally higher than α-tubulins39,48, also confirmed in tardigrades. Notably, tubulin sequences from Ech. testudo and Milnesium t. tardigradum Doyère, 1840 (newly Milnesium inceptum Morek, Suzuki, Schill, Georgiev, Yankova, Marley & Michalczyk, 201949) were mostly truncated, which might reflect the poorer quality of the available sequencing data. Other tardigrade tubulin proteins could still be found with improved transcriptomic, genomic, and proteomic approaches in the future, especially in the abovementioned species.
We found three tardigrade α-tubulin isoforms: Tub1A1, Tub1A2 and Tub1C1. The Tub1C1 was likely present in the ancient Protostomia since its group contains a mollusk Tub1C. The Tub1C1 might have gotten lost in heterotardigrades; however, fragmented and truncated tubulin sequences found in analyzed heterotardigrade transcriptomes make it hard to draw a more definitive conclusion. The Tub1A1 and Tub1A2 form a sister group to Tub1C1 and seem present only in tardigrades. This cluster comprises both Hetero- and Eutardigrada sequences indicating that these isoforms arose by duplication in the last common tardigrade ancestor.
The isoform Tub1A1 has no C-terminal tyrosine residue—its C-terminal AA motif is "EEV." The Tub1C1 possesses a C-terminal tyrosine or rarely phenylalanine; however, it lacks the conserved predeceasing double glutamate—its C-terminal AA motif is "NGEY/F." The question is whether Tub1A1 and Tub1C1 can undergo the classical tyrosination and detyrosination cycle that is typical for most of α-tubulins across many species29. On the other hand, all found tardigrade α-tubulins possess a lysine residue equivalent to vertebrate Lys40 that is a target for acetylation29. Indeed, an anti-acetylated tubulin antibody was successfully used in studies on tardigrade nervous system21,31.
Tardigrade β-tubulin sequences formed seven clusters, many reflecting the class or order distinction of respective taxa. The Tub2D2s from the order Apochela (Tub2D2-Apo) were separated from the other eutardigrades (Tub2D2-Eu). The Tub2D4 sequences were split based on the membership to the class Eutardigrada or Heterotardigrada, in Tub2D4-Eu and Tub2D4-Het, respectively. The Tub2D3, Tub2C and Tub2A formed independent clusters, with the Tub2A being the most distant. Although all sequences in this cluster are truncated, their sequence signature is distinct from the other clusters, justifying the existence of this Ech. testudo-specific isoform. C-termini of tardigrade β-tubulins are more diverse compared to α-tubulins, and finding some conserved sequences was difficult.
An interesting isoform is the Tub2C, which might be a homolog of vertebrate βIII-tubulins expressed primarily in neurons and in certain cancer cells42. We base our hypothesis on the presence of a hydrophobic AA at the relative position 371 and an alanine 295 in our alignment (designated as AA351 and AA275, respectively, in the literature on vertebrate tubulins42). The change from a polar threonine to a non-polar isoleucine or valine at the position equivalent to the vertebrate AA351 is conserved in many βIII-tubulins in vertebrates and even invertebrates42,43. Interestingly, the highly extremotolerant Ramazzottius varieornatus Bertolani & Kinchin, 199350 lacks the Tub2C isoform and the ε-tubulin as well51. These findings imply that the function of Tub2C must be investigated directly in tardigrades in the future.
We wanted to isolate the CDS of tardigrade tubulins directly from tardigrades and test them in cells to verify their function. All tardigrade tubulins tagged with fluorescent proteins (mScarlet or mEGFP) correctly localized to predicted subcellular locations in mammalian cells. All β-tubulins were well incorporated into mammalian microtubules, and their presence was always easily detectable. On the contrary, tardigrade α-tubulins localized to microtubules relatively poorly. The best incorporation was detected for Tub1A1, and the worst microtubule incorporation was observed for Tub1A2. This tubulin exhibited mostly cytoplasmic localization; however, some microtubular localization was also detectable. The Tub1C1 also appeared frequently cytoplasmic; however, microtubule localization became apparent after a longer expression (> 48 h).
Differences in the ability of mEGFP-tagged α-tubulins to incorporate into mammalian microtubules were surprising. It should be noted that the rest of the plasmid backbone, including the linker between the tubulin CDS and the fluorescent protein CDS, was identical. One of the possible explanations for the differences might be the heterogeneity of C-termini, which might impact tubulin heterodimer assembly or incorporation into microtubules. Since we expressed tardigrade tubulins in mammalian cells, an aberrant interaction with some tubulin interactors might have occurred, for instance, with the endogenous tubulin tyrosine ligase (TTL)52,53, influencing the incorporation of tardigrade α-tubulins—especially Tub1A2—to microtubules.
The different abilities of individual tardigrade α-tubulins to incorporate into microtubules could also be influenced by temperature. The optimal temperature for Hys. exemplaris is between 10 and 18 °C and temperatures above 22 °C are already critical for survival54. In contrast, the optimal temperature for mammalian cells used here is 37 °C. Generally, suboptimal temperature causes a decrease in rates of microtubule polymerization and depolymerization, the amount of polymer assembled before catastrophes, and the frequency of microtubule nucleation55. However, it is interesting that tardigrade β-tubulins were unaffected, and this phenomenon warrants further investigation, ideally directly in tardigrades.
The ε-tubulin has been identified in all eukaryotic kingdoms but has been lost independently in many clades several times39. Remarkably, our data do not support the "ZED module" hypothesis stating that: "(1) organisms that lack epsilon-tubulin always also lack delta- and zeta-tubulin and (2) organisms that have epsilon-tubulin always also have delta- and/or zeta-tubulin56." Notably, the ZED module hypothesis was based on a much smaller dataset than that of Findeisen et al.39, which we used as a reference in our study.
Currently, tardigrades are classified in the clade Panarthropoda, a part of the superphylum Ecdysozoa. However, the phylogenetic position of tardigrades within the Ecdysozoa remains controversial and debated20,21,22,23,24,57. Although tubulins are generally unsuitable for inference of species' phylogenetic position due to the high degree of tubulin gene duplications—especially α- and β-tubulin—occurring spontaneously and independently throughout the evolution39, our analysis might still contribute to the refinement of the phylogenetic position of Tardigrada. All Ecdysozoa lack ζ-tubulin39. Remarkably, Nematoda also lost their δ- and ε-tubulins; however, some groups of Arthropoda still possess them, including Hymenoptera39. We found three unique tardigrade ε-tubulin sequences, two in heterotardigrades and one in the eutardigrade Hys. exemplaris. Hence, our data indicate that the current placement of tardigrades among Panarthropoda is correct.
Interestingly, δ-, ε- or ζ-tubulins are not essential for functional cilia in general since there are ciliated species lacking all these isotypes39. However, these tubulins are found in species that possess cilia, where they play roles in ciliary assembly and function39,56,58,59. Indeed, tardigrades possess cilia that fulfill important sensory tasks2. Investigation of the role of tardigrade ε-tubulin for tardigrade ciliary functions will be an exciting avenue of future research. Moreover, our work opens the way for exploration of the tardigrade tubulin code29,60 and its role in the fascinating survival abilities of tardigrades.
Methods
Identification of tardigrade tubulin sequences
We collected tubulin protein sequences of four model organisms Homo sapiens, Drosophila melanogaster, C. elegans and Mus musculus from NCBI. We also acquired published genomes and transcriptomes of eight tardigrade taxa—Ecn. cf. sigismundi61, Richtersius cf. coronifer61, Ech. testudo62, Mesobiotus philippinicus Mapalo, Stec, Mirano-Bascos & Michalczyk, 2016 62,63, Milnesium tardigradum (Genbank GFGZ00000000.1), Pam. richtersi (Genbank GFGY00000000.1), Hys. dujardini (Doyère, 1840) (now Hys. exemplaris) (Hys. dujardini strain: Z151 Genome sequencing and assembly; (Yoshida et al.24) and Ram. varieornatus (Ram. varieornatus genome, (Hashimoto et al.51) (Table 1). Of note, originally, we used genome assemblies of those last two strains from the website tardigrades.org, which has been discontinued. Regarding name abbreviations of tardigrade species/taxa, we followed Perry et al.64.
By using the BLAST + software (Version 2.2.26+)65, we prepared our local BLAST + database from the abovementioned data. We ran a TBLASTN search in the local tardigrade BLAST + database with an e-value cutoff of 1E10–5 using tubulin protein sequences from H. sapiens, M. musculus, D. melanogaster and C. elegans as a query (Supplementary Data 5). Multiplicate redundant sequences were filtered out based on unique sequence IDs. Hits with a unique sequence ID were further aligned using Clustal Omega66 and identical redundant sequences from the same species were removed. The resulting dataset of about 90 putative tardigrade tubulin sequences was transferred to Benchling and translated based on coordinates from the BLAST + search (frame and strand) (UJEP dataset).
Alternatively, the unique sequences were translated using the esl-translate function of HMMER package (3.1b2 February 2015) and an in-house macro for trimming erroneous N-terminal AA extensions arising from artificial translation of 5' segments of identified transcripts (CRG dataset). To extract the domain sequences of the tardigrade tubulin proteins, we used hmmscan from the HMMER package with the default parameters to scan Pfam32.0 67 and retrieved the hits corresponding to the Pfam tubulin family (PF00091.25) (CRG tubulin domains dataset).
Multiple Sequence Alignments computation and phylogenetic inference
The phylogenetic analysis was carried out in two steps. First, we computed multiple sequence alignments of the found putative tardigrade tubulin protein or domain sequences together with a previously published large tubulin protein dataset39 relying on the regressive mode of TCoffee38 (Version_13.45.58.c355d11 with Clustal Omega66 guide trees and MAFFT-G-INS-i alignments68).
Second, the produced alignments were used as input for phylogenetic reconstruction tools based on maximum likelihood (IQ-TREE69, version 2.1.2) and minimum evolution (FastME70, version 2.1.6.1).
Culture of Hys. exemplaris
Samples of Hys. exemplaris were obtained from envirocom (Tübingen, Germany) and from our own culture. Eutardigrade Hys. exemplaris is a freshwater, herbivorous species that typically feeds on algae and was cultured as described previously54, but with small modifications. Briefly, tardigrades were cultured on 90 mm plastic Petri dishes. The bottom of all dishes was scratched with a fine sandpaper to allow easier locomotion of tardigrades. Each dish contained 10 mL of Chalkley's medium (0.004 g/L KCl, 0.1 g/L NaCl, 0.006 g/L CaCl2, 20 ml/L soil extract; Soil extract was an autoclaved supernatant of fertile, humus-enriched soil and distilled water in 1:2 ratio. The extract was allowed to settle over a few days and then filtered) mixed with 50 mL of spring water (Rajec, Czech Republic). Cultures were fed with green algae Chlorococcum sp. by adding ca. one volume concentrated algal culture to four volumes of the tardigrade culture. Each culture was checked every 2 to 3 days with a microscope to ensure tardigrades flourished and the oxygen supply was provided by pipetting. According to need, tardigrades were fed the algae every one or two weeks. During the cleaning/checking procedures, a thin iridescent film on the surface of the medium, dead animals, old exuviae, and different organic residues were removed using a pipette. The 3⁄4 volume of the medium were changed once every ten days. This approach has proven successful in that not all food was consumed and the medium was still fresh. In each Petri dish, the population numbered around 500 individuals. If the number of specimens became too large, juveniles and eggs were transferred to a new culture dish. Cultures were stored in a Q-Cell incubator equipped with a combined cooling and heating system with forced air circulation. The temperature was constantly set at 18.7 °C. The external light source (INVITAL LED) with a combination of white and blue LED was installed into the incubator with a 10 h/14 h (light/dark) photoperiod. Algal culture media have several components in common—sources of nitrogen, phosphorus, vitamins and trace metals. For the culture of Chlorococcum sp., we used Bold’s Basal Medium71 as a basis. The algae were grown in 250 mL Erlenmeyer flasks with the mouth covered by two layers of parafilm. The algae settled at the bottom, therefore, they were shaken by hand once a week. Flasks were located in the laboratory out of the direct sunlight. The laboratory room temperatures fluctuated between 20 and 24 °C.
Sample collection and mRNA isolation from Hys. exemplaris specimens
We collected adult specimens (ca. 200 μm to 300 μm in length) in their culture medium and removed various contaminating substances using a pipette. Specimens were washed several times, first with distilled water and then with ultrapure water. Tardigrades (ca. 2000 individuals) were placed in a low-retention 1.5 mL microcentrifuge tube. The tube was centrifuged at 10,000 × g for 1 min at room temperature. During centrifugation, tardigrades sedimented at the bottom of the tube and compacted into a visible pellet. The supernatant was carefully removed as much as possible without disrupting the pellet. In some cases, samples were flash frozen in liquid nitrogen at this stage. Otherwise, we were quickly crushing the pellet with a sterile plastic pestle for 3 s at room temperature and then resuspend it by adding 200 μL of RLT buffer from the RNeasy Mini Kit (74104, Qiagen) that contained 1% (v/v) 2-mercaptoethanol. Then, we crushed the sample again for 5 s and rapidly froze it in liquid nitrogen. As a next step, we removed the tube from the liquid nitrogen and allowed the sample to thaw. As the sample started to thaw, we were crushing it again for 10 s. We repeated the freeze/thaw/crushing steps five times (until homogenization). At the end, we washed the pestle with 150 μL RLT buffer with 2-mercaptoethanol. For sample homogenization, we mixed the sample five times with a 20 G syringe. We used RNeasy Mini Kit for total RNA isolation according to manufacturer's instructions. The RNA was eluted with RNase-free water. The concentration was measured by UV/VIS spectrophotometer Denovix. RNA was stored at − 80 °C.
Reverse transcription
The RNA template was converted into cDNA by SuperScript® IV Reverse Transcriptase (18090010, Thermo Fisher Scientific) using oligo dT primers according to manufacturer's instructions. We used RNaseH (M0297S, NEB) to remove RNA duplexed with the newly synthetized cDNA. The cDNA was stored at − 20 °C.
Cloning of tardigrade tubulin coding sequences
Individual tardigrade tubulin CDS were isolated by PCR using specific forward (fwd) and reverse (rev) primers (Supplementary Table S3) and cDNA from adult Hys. exemplaris specimens as a template. The PCR products were purified on an agarose gel and cloned into the pJet1.2 (CloneJet kit, K1231, Thermo Fisher Scientific). To tag tardigrade tubulin CDS with fluorescent proteins, we employed NEBuilder® HiFi DNA Assembly Cloning Kit (E2621L, NEB).
Tardigrade β-tubulins and γ-tubulin were tagged with mScarlet. Corresponding tubulin CDS were amplified from above described pJet1.2-tubulin vectors by PCR using specific primers (Supplementary Table S3, designated as "HiFi"). The pLifeAct_mScarlet-i_N1 (a gift from Dorus Gadella72; Addgene plasmid # 85,056; http://n2t.net/addgene:85056; RRID:Addgene_85056) was linearized by NheI and BamHI, which led to the removal of the LifeAct CDS. The linearized recipient plasmid and individual tubulin CDS were assembled using HiFi DNA Assembly creating fusion CDS of tubulins tagged on their C-terminus with mScarlet. The newly characterized Hys. exemplaris γ-tubulin obtained Genbank accession number OQ134936.
Tardigrade α-tubulins and ε-tubulin were tagged with mEGFP. First, mEGFP was amplified by PCR using specific primers (Supplementary Table S3, designated as "HiFi") for each tubulin CDS from pGEMHE-NLS-mEGFP (a gift from Melina Schuh73; Addgene plasmid # 105,527; http://n2t.net/addgene:105527; RRID:Addgene_105527). Second, each tubulin CDS was amplified by PCR using specific primers (Supplementary Table S3, designated as "HiFi") from above described pJet1.2-tubulin vectors. Third, pEGFP-C1 (Clontech) was linearized by NheI and HindIII which resulted in the removal of EGFP CDS. Finally, recipient plasmid backbone, mEGFP CDS and corresponding tubulin CDS were assembled using HiFi DNA Assembly creating fusion CDS of tubulins tagged on their N-terminus with mEGFP. All plasmids were verified by sequencing. pEGFP-Tub (encoding EGFP-TUBA1B) was originally from Clontech.
Transfection and confocal microscopy
The hTert-RPE1 and U87-MG cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) or Eagle’s minimal essential medium (E-MEM), respectively, supplemented with 10% (v/v) fetal bovine serum and 0.1% (w/v) penicillin and 0.1% (w/v) streptomycin. Cells were maintained in culture flasks (Falcon or Corning) at 37 °C and 5% CO2 in a humid atmosphere in an incubator and subcultured (passaged) every 2 to 3 days after obtaining 80% to 90% confluence. Cells were seeded on 18-well glass-bottom dishes (ibidi) one day before transfection and then transfected using Lipofectamine 3000 (Thermo) or JetOptimus (Polyplus) according to manufacturer’s instructions. Cells were incubated in the incubator and visualized 24 h to 48 h post transfection.
Transfected cells were imaged using a Leica SP8 confocal microscope enclosed in an environmental chamber with constant temperature (36.9 °C), humidity (95%) and 5% CO2 levels. Images were acquired using a 63x/1.40 N.A. oil objective. Filter and detector settings were optimised to minimise any possible bleed-through between fluorescence channels.
Fluorescence images were adjusted in Fiji74. All acquired z-stack were processed using Maximum intensity projection and Brightness/Contrast function. Only the Tub1C1 was additionally processed using Substract Background function with 150 pixels rolling ball radius.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author (stanislav.vinopal@ujep.cz). Recombinant DNA generated in this study is available upon request from the corresponding author. The datasets generated and/or analysed during the current study are included in this published article (and its Supplementary Information files) and are also available in the repository Zenodo (https://doi.org/10.5281/zenodo.7528263). All sequenced tubulin CDS of Hys. exemplaris were uploaded to GenBank (tHe-Tub1A1, OQ282841; tHe-Tub1A2, OQ282842; tHe-Tub1C1, OQ282843; tHe-Tub2C, OQ282844; tHe-Tub2D2, OQ282845; tHe-Tub2D3, OQ282846; tHe-Tub2D4, OQ282847; tHe-Tub3, OQ134936; tHe-Tub5, OQ282848).
References
Fleming, J. F. & Arakawa, K. Systematics of tardigrada: A reanalysis of tardigrade taxonomy with specific reference to Guil et al. (2019). Zool. Scr. 50, 376–382 (2021).
Møbjerg, N., Jørgensen, A., Kristensen, R. M. & Neves, R. C. Morphology and functional anatomy. In Water Bears: The Biology of Tardigrades (ed. Schill, R. O.) 57–94 (Springer International Publishing, 2018).
Halberg, K. A., Jørgensen, A. & Møbjerg, N. Desiccation tolerance in the tardigrade Richtersius coronifer relies on muscle mediated structural reorganization. PLoS ONE 8, e85091 (2013).
Czerneková, M., Jönsson, K. I., Chajec, L., Student, S. & Poprawa, I. The structure of the desiccated Richtersius coronifer (Richters, 1903). Protoplasma 254, 1367–1377 (2017).
Czerneková, M. & Vinopal, S. The tardigrade cuticle. Limnol. Rev. 21, 127–146 (2021).
Schill, R. O. & Hengherr, S. Environmental adaptations: Desiccation tolerance. In Water Bears: The Biology of Tardigrades (ed. Schill, R. O.) 273–293 (Springer International Publishing, 2018).
Jönsson, K. I., Borsari, S. & Rebecchi, L. Anhydrobiotic survival in populations of the tardigrades Richtersius coronifer and Ramazzottius oberhaeuseri from Italy and Sweden. Zool. Anzeiger A J. Comp. Zool. 240, 419–423 (2001).
Rebecchi, L., Altiero, T. & Guidetti, R. Anhydrobiosis: the extreme limit of desiccation tolerance. Invertebr. Surv. J. 4, 65–81 (2007).
Schill, R. O. & Fritz, G. B. Desiccation tolerance in embryonic stages of the tardigrade. J. Zool. 276, 103–107 (2008).
Wełnicz, W., Grohme, M. A., Kaczmarek, Ł, Schill, R. O. & Frohme, M. Anhydrobiosis in tardigrades—The last decade. J. Insect Physiol. 57, 577–583 (2011).
Czernekova, M. & Jönsson, K. I. Experimentally induced repeated anhydrobiosis in the eutardigrade Richtersius coronifer. PLoS ONE 11, e0164062 (2016).
Guidetti, R., Rizzo, A. M., Altiero, T. & Rebecchi, L. What can we learn from the toughest animals of the Earth? Water bears (tardigrades) as multicellular model organisms in order to perform scientific preparations for lunar exploration. Planet. Space Sci. 74, 97–102 (2012).
Kaczmarek, Ł et al. Staying young and fit? Ontogenetic and phylogenetic consequences of animal anhydrobiosis. J. Zool. 309, 1–11 (2019).
Hengherr, S., Brümmer, F. & Schill, R. O. Anhydrobiosis in tardigrades and its effects on longevity traits. J. Zool. 275, 216–220 (2008).
Arakawa, K. Examples of extreme survival: tardigrade genomics and molecular anhydrobiology. Annu. Rev. Anim. Biosci. 10, 17–37 (2022).
Hesgrove, C. & Boothby, T. C. The biology of tardigrade disordered proteins in extreme stress tolerance. Cell Commun. Signal. 18, 178 (2020).
Yagi-Utsumi, M. et al. Desiccation-induced fibrous condensation of CAHS protein from an anhydrobiotic tardigrade. Sci. Rep. 11, 21328 (2021).
Tanaka, A. et al. Stress-dependent cell stiffening by tardigrade tolerance proteins that reversibly form a filamentous network and gel. PLOS Biol. 20, e3001780 (2022).
Kasianchuk, N., Rzymski, P. & Kaczmarek, Ł. The biomedical potential of tardigrade proteins: A review. Biomed. Pharmacother. 158, 114063 (2023).
Jørgensen, A., Kristensen, R. M. & Møbjerg, N. Phylogeny and Integrative Taxonomy of Tardigrada. In Water Bears: The Biology of Tardigrades (ed. Schill, R. O.) 95–114 (Springer International Publishing, 2018).
Mayer, G. et al. Selective neuronal staining in tardigrades and onychophorans provides insights into the evolution of segmental ganglia in panarthropods. BMC Evol. Biol. 13, 230 (2013).
Rota-Stabelli, O. et al. Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda. Genome Biol. Evol. 2, 425–440 (2010).
Rota-Stabelli, O., Daley, A. C. & Pisani, D. Molecular timetrees reveal a cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr. Biol. 23, 392–398 (2013).
Yoshida, Y. et al. Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLOS Biol. 15, e2002266 (2017).
Pollard, T. D. & Goldman, R. D. Overview of the cytoskeleton from an evolutionary perspective. Cold Spring Harb. Perspect. Biol. 10, a030288 (2018).
Herrmann, H. & Strelkov, S. V. History and phylogeny of intermediate filaments: Now in insects. BMC Biol. 9, 16 (2011).
Goldstein, L. S. B. & Gunawardena, S. Flying through the Drosophila cytoskeletal genome. J. Cell Biol. 150, F63–F68 (2000).
Hering, L., Bouameur, J.-E.E., Reichelt, J., Magin, T. M. & Mayer, G. Novel origin of lamin-derived cytoplasmic intermediate filaments in tardigrades. Elife 5, 1–18 (2016).
Janke, C. & Magiera, M. M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 21, 307–326 (2020).
Walz, B. Molting in Tardigrada. A review including new results on cuticle formation in Macrobiotus hufelandi. In Proceedings of the Third International Symposium on the Tardigrada (ed. Nelson, D. R.) 129–142 (East Tennessee State University Press, 1982).
Gross, V. & Mayer, G. Neural development in the tardigrade Hypsibius dujardini based on anti-acetylated α-tubulin immunolabeling. EvoDevo 6, 12 (2015).
Mayer, G., Kauschke, S., Rüdiger, J. & Stevenson, P. A. Neural markers reveal a one-segmented head in tardigrades (water bears). PLoS ONE 8, e59090 (2013).
Persson, D. K., Halberg, K. A., Jørgensen, A., Møbjerg, N. & Kristensen, R. M. Neuroanatomy of Halobiotus crispae (Eutardigrada: Hypsibiidae): Tardigrade brain structure supports the clade panarthropoda. J. Morphol. 273, 1227–1245 (2012).
Persson, D. K., Halberg, K. A., Jørgensen, A., Møbjerg, N. & Kristensen, R. M. Brain anatomy of the marine tardigrade Actinarctus doryphorus (Arthrotardigrada). J. Morphol. 275, 173–190 (2014).
Smith, F. W. & Jockusch, E. L. The metameric pattern of Hypsibius dujardini (Eutardigrada) and its relationship to that of other panarthropods. Front. Zool. 11, 66 (2014).
Schulze, C., Neves, R. C. & Schmidt-Rhaesa, A. Comparative immunohistochemical investigation on the nervous system of two species of Arthrotardigrada (Heterotardigrada, Tardigrada). Zool. Anz. 253, 225–235 (2014).
Zantke, J., Wolff, C. & Scholtz, G. Three-dimensional reconstruction of the central nervous system of Macrobiotus hufelandi (Eutardigrada, Parachela): implications for the phylogenetic position of Tardigrada. Zoomorphology 127, 21–36 (2008).
Garriga, E. et al. Large multiple sequence alignments with a root-to-leaf regressive method. Nat. Biotechnol. 37, 1466–1470 (2019).
Findeisen, P. et al. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol. Evol. 6, 2274–2288 (2014).
Luallen, R. J. et al. Discovery of a natural microsporidian pathogen with a broad tissue tropism in Caenorhabditis elegans. PLOS Pathog. 12, e1005724 (2016).
Gąsiorek, P. & Kristensen, R. M. New marine heterotardigrade lineages (Echiniscoididae) from the tropics. Eur. Zool. J. 89, 719–754 (2022).
Ludueña, R. F. Possible roles of specific amino acids in β-tubulin isotypes in the growth and maintenance of neurons: novel insights from cephalopod mollusks. Front. Mol. Neurosci. 15, 838393 (2022).
Wang, W. et al. Novel mutations involving βI-, βIIA-, or βIVB-tubulin isotypes with functional resemblance to βIII-tubulin in breast cancer. Protoplasma 254, 1163–1173 (2017).
Gąsiorek, P., Stec, D., Morek, W. & Michalczyk, Ł. An integrative redescription of Hypsibius dujardini (Doyère, 1840), the nominal taxon for Hypsibioidea (Tardigrada: Eutardigrada). Zootaxa 4415, 45 (2018).
Brunak, S., Engelbrecht, J. & Knudsen, S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 220, 49–65 (1991).
Hebsgaard, S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452 (1996).
Yutin, N. & Koonin, E. V. Archaeal origin of tubulin. Biol. Direct 7, 10 (2012).
Zhao, Z. et al. Molecular evolution and functional divergence of tubulin superfamily in the fungal tree of life. Sci. Rep. 4, 6746 (2015).
Morek, W. et al. Redescription of Milnesium alpigenum Ehrenberg, 1853 (Tardigrada: Apochela) and a description of Milnesium inceptum sp. nov., a tardigrade laboratory model. Zootaxa 4586, 35–64 (2019).
Bertolani, R. & Kinchin, I. M. A new species of Ramazzottius (Tardigrada, Hypsibiidae) in a rain gutter sediment from England. Zool. J. Linn. Soc. 109, 327–333 (1993).
Hashimoto, T. et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat. Commun. 7, 12808 (2016).
Knossow, M., Campanacci, V., Khodja, L. A. & Gigant, B. The mechanism of tubulin assembly into microtubules: Insights from structural studies. iScience 23, 101511 (2020).
Szyk, A., Deaconescu, A. M., Piszczek, G. & Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18, 1250–1258 (2011).
Roszkowska, M. et al. Tips and tricks how to culture water bears: Simple protocols for culturing eutardigrades (Tardigrada) under laboratory conditions. Eur. Zool. J. 88, 449–465 (2021).
Li, G. & Moore, J. K. Microtubule dynamics at low temperature: Evidence that tubulin recycling limits assembly. Mol. Biol. Cell 31, 1154–1166 (2020).
Turk, E. et al. Zeta-tubulin is a member of a conserved tubulin module and is a component of the centriolar basal foot in multiciliated cells. Curr. Biol. 25, 2177–2183 (2015).
Smith, F. W., Bartels, P. J. & Goldstein, B. A hypothesis for the composition of the tardigrade brain and its implications for panarthropod brain evolution. Integr. Comp. Biol. 57, 546–559 (2017).
Stathatos, G. G., Dunleavy, J. E. M., Zenker, J. & O’Bryan, M. K. Delta and epsilon tubulin in mammalian development. Trends Cell Biol. 31, 774–787 (2021).
Chang, P., Giddings, T. H., Winey, M. & Stearns, T. ɛ-Tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 5, 71–76 (2003).
Roll-Mecak, A. The tubulin code in microtubule dynamics and information encoding. Dev. Cell 54, 7–20 (2020).
Kamilari, M., Jørgensen, A., Schiøtt, M. & Møbjerg, N. Comparative transcriptomics suggest unique molecular adaptations within tardigrade lineages. BMC Genomics 20, 607 (2019).
Mapalo, M. A. et al. The unique antimicrobial recognition and signaling pathways in tardigrades with a comparison across ecdysozoa. Genes Genomes Genet. 10, 1137–1148 (2020).
Mapalo, M. A., Stec, D., Mirano-Bafscos, D. & Michalczyk, Ł. Mesobiotus philippinicus sp. nov., the first limnoterrestrial tardigrade from the Philippines. Zootaxa 4126, 411 (2016).
Perry, E., Miller, W. R. & Kaczmarek, Ł. Recommended abbreviations for the names of genera of the phylum Tardigrada. Zootaxa 4608, 145 (2019).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Lefort, V., Desper, R. & Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800 (2015).
Littler, D. S., Hellebust, J. A., Litter, M. M. & Craigie, J. S. Handbook of Phycological Methods: Culture Methods and Growth Measurements (Cambridge University Press, 1973).
Bindels, D. S. et al. MScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56 (2016).
Clift, D. et al. A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692-1706.e18 (2017).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
The project was supported by a Grant No.: UJEP-SGS-2021-53-003-2 within the Student grant competition at UJEP (K.N.F., M.C., J.S., S.V.). The authors acknowledge the assistance provided by the Research Infrastructure NanoEnviCz (Project No. LM2018124) and the project Pro-NanoEnviCz (Reg. No. CZ.02.1.01/0.0/0.0/16_013/0001821 and CZ.02.1.01/0.0/0.0/18_046/0015586), supported by the Ministry of Education, Youth and Sports of the Czech Republic and the European Union European Structural and Investments Funds in the frame of the Operational Programme Research Development and Education (K.N.F., M.C., J.S., J.M., S.V.). Studies have been conducted in the framework of activities of BARg (Biodiversity and Astrobiology Research group, Ł.K.). The research leading to these results has also received funding from the Spanish Ministry of Science and Innovation (A.B and C.N.). We acknowledge the support of the Spanish Ministry of Science and Innovation to the EMBL partnership, to Centro de Excelencia Severo Ochoa and to the CERCA Programme of Generalitat de Catalunya (A.B and C.N.).
Author information
Authors and Affiliations
Contributions
Conceptualization, S.V.; Methodology, K.N.F., A.B., M.C., J.S., S.V.; Investigation, K.N.F., A.B., S.V.; Resources, S.V., C.N., M.C., Ł.K., J.M.; Formal analysis, K.N.F., A.B., J.S., S.V.; Writing Original Draft, S.V., Writing – Review & Editing, K.N.F., A.B., M.C., J.S., C.N., Ł.K., J.M., S.V.; Funding acquisition, C.N., Ł.K., J.M., S.V.; Supervision, C.N., J.M., Ł.K., S.V.; Supervision of the project, S.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Novotná Floriančičová, K., Baltzis, A., Smejkal, J. et al. Phylogenetic and functional characterization of water bears (Tardigrada) tubulins. Sci Rep 13, 5194 (2023). https://doi.org/10.1038/s41598-023-31992-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-31992-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.