Abstract
Notch1 plays important roles in T cell development and is highly expressed in activated CD4+ T cells. However, the underlying mechanism of Notch1 transcription in T cells has not been fully characterized. Therefore, we aimed to determine how Notch1 expression is regulated during the activation of CD4+ T cells. Both the surface expression and mRNA transcription of Notch1 were significantly higher in activated CD4+ T cells, but the inhibition of phosphatidylinositol 3-kinase (PI3K) by LY294002 or deletion of the Pdk1 gene impaired this upregulation of Notch1. Interrogation of the Notch1 promoter region using serially deleted Notch1 promoter reporters revealed that the − 300 to − 270 region is crucial for its transcription in activated T cells. In addition, we found that nuclear factor (NF)-κB subunits containing RelA bind directly to this promoter region, thereby upregulating transcription. In addition, inhibition of NF-κB by SN50 impaired upregulation of Notch1 surface protein and mRNA in activated CD4+ T cells. Thus, we provide evidence that Notch1 transcription in activated CD4+ T cells is upregulated via the PI3K-PDK1-NF-κB signaling pathway.
Similar content being viewed by others
Introduction
The Notch family comprises four conserved transmembrane receptors (Notch 1–4) that contain an ectodomain with epidermal growth factor-like repeats; an intracellular domain containing a regulation of amino acid metabolism (RAM) 23 domain, ankyrin repeats, and a proline, glutamate, serine, threonine-rich (PEST) domain1. These are single-pass transmembrane heterodimeric receptors, in which the N-terminal extracellular subunit and the C-terminal intracellular subunit, which includes a transmembrane domain, are noncovalently associated2. Notch signaling is triggered by the binding of ligands, which include Jagged 1, Jagged 2, Delta-like 1, Delta-like 3, and Delta-like 43,4,5. When a ligand binds to Notch1, the extracellular domain of Notch1 is cleaved by the ADAM10 metalloprotease, and subsequent proteolytic cleavage occurs in the intracellular domain as a result of activity of the γ-secretase complex6,7,8. The released Notch1 intracellular domain then translocates to the nucleus, where it regulates transcription6. Specifically, the cleaved Notch1 intracellular domain interacts with a recombination signal binding protein for immunoglobulin kappa J region (RBP-Jκ), thereby converting it from a transcriptional repressor to an activator, which results in target gene expression9.
Notch1 plays a key role in T cell development and lineage commitment. Hematopoietic stem cell-derived progenitors from the bone marrow have the potential to become either T or B cells10. Studies of the overexpression of the active form and knockout of Notch1 have shown that Notch1 signaling affects whether progenitor cells are directed down the T or B cell lineage, promoting T cell, rather than B cell, differentiation11,12. After commitment to the T cell lineage, the T cell progenitor cells differentiate into αβ or γδ T cells. Previous studies using mouse model systems suggest that a reduction in Notch1 signaling increases differentiation of γδ T cells at the expense of αβ T cells13,14. However, the opposite effect was found in human cells15. In addition, Notch signaling regulates various aspects of T cell function, including their activation, differentiation, expansion, and cytokine production9. Furthermore, recent studies of T cell acute lymphoblastic leukemia (T-ALL) have revealed that an activating mutation in Notch1 commonly causes leukemia in cooperation with other oncogenic mutations16. Therefore, Notch1 has emerged as an important therapeutic target for T-ALL.
Despite the importance of Notch1 in T cells, it remains unclear how Notch1 expression is regulated in this cell type. There have been some previous studies of the transcriptional regulation of Notch1 in non-T cells. p53 is a well-known transcriptional regulator of Notch1 in keratinocytes, in which it activates transcription by directly binding to the promoter region17,18. Kruppel-like factor (Klf) 4 and Sp3 are other transcriptional regulators of Notch1 in keratinocytes, in which they bind to the Notch1 promoter, causing a reduction in gene expression19. Recent studies show that distal-less homeobox (DLX) 5 binds directly to the Notch1 promoter, which triggers upregulation of gene transcription in osteosarcoma cells, and that SIRT6 epigenetically regulates Notch1 transcription via DNA methyltransferase (DNMT) 1 in adipose-derived stem cells20,21. However, their relevance for Notch1 transcription in T cells has not been evaluated. Therefore, we aimed to determine how Notch1 expression is regulated in T cells by attempting to identify the most important promoter region for Notch1 transcription and the key transcriptional regulators. To this end, we used serial deletions of the Notch1 promoter region and found that the − 300 to − 270 region is critical for the transcription of the gene in CD4+ T cells. In addition, we found that the RelA-containing nuclear factor (NF)-κB complex proteins directly bind to this region to positively regulate transcription.
Results
Notch1 expression is induced by phosphoinositide 3-phosphate (PI3K)-phosphoinositide-dependent kinase-1 (PDK1) signaling
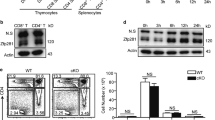
We first determined whether Notch1 expression is regulated during T cell activation. Negatively isolated primary CD4+ T cells were activated using anti-CD3 and anti-CD28 antibodies, and flow cytometric analysis showed that the surface expression of Notch1 was much higher after activation (Fig. 1a, b). We next determined whether the high Notch1 surface expression was the result of upregulation of Notch1 transcription, and found that Notch1 mRNA expression increased in a time-dependent manner after activation using anti-CD3 and anti-CD28 antibodies (Fig. 1c). These findings imply that the high surface expression of Notch1 is principally the result of the transcriptional activation of Notch1. Interestingly, the activation-induced Notch1 expression was abolished by treatment with LY294002, a PI3K inhibitor. Over half of the activated CD4+ T cells showed an increase in surface Notch1 expression after 72 h, but almost none of the LY294002-treated CD4+ T cells upregulated Notch1 (Fig. 1d, e), which implies that the PI3K pathway plays a critical role in Notch1 expression. In addition, expression of Notch1 mRNA levels fell markedly in activated CD4+ T cells treated with LY294002 (Fig. 1f). We also checked whether reduced expression of Notch1 surface protein and mRNA is due to destabilization of Notch1 mRNA after treatment with LY294002; however, LY294002 treatment did not accelerate decay of Notch1 mRNA (Supplementary Fig. 1). Because PDK1 is a principal downstream effector of PI3K, we next measured Notch1 expression in primary CD4+ T cells isolated from mice with CD4+ T cell-specific deletion of PDK1 that were activated using anti-CD3 and anti-CD28 antibodies. We found that upregulation of Notch1 expression was impaired markedly in Pdk1-deficient CD4+ T cells (Fig. 1g, h). Thus, PI3K-PDK1 signaling increases Notch1 expression through a robust induction of transcription in activated CD4+ T cells.
Notch1 expression is upregulated in activated primary CD4+ T cells via the PI3K-PDK1 pathway. (a, b) Flow cytometry for surface Notch1 expression after cellular activation using anti-CD3 and anti-CD28 antibodies for the indicated times. (c) Quantitative real-time PCR for Notch1 mRNA expression after cellular activation. (d, e) Flow cytometry for surface Notch1 expression after treatment with the PI3K inhibitor LY294002. (f) Quantitative real-time PCR to detect Notch1 mRNA after cells were activated by anti-CD3 and anti-CD28 antibodies for 48 h in the presence of LY294002. (g, h) Flow cytometry for surface Notch1 expression in PDK1-deficient CD4+ T cells. Data are mean ± SD. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 (Student’s t-test).
The − 300 to − 270 region of Notch1 is crucial for its transcription
The upstream region (up to − 400 base pairs) from the start codon of Notch1 is highly conserved among vertebrates; this conserved region continues intermittently until − 1000 base pairs (Fig. 2a). To identify the essential cis element for Notch1 transcriptional regulation, sections of the Notch1 promoter region (+ 1 to − 1140) were serially deleted, and the deleted fragments were cloned into the pGL3 basic reporter plasmid (Fig. 2b). These vectors were transfected into Jurkat T cells, the cells were activated using PMA/ionomycin, and a luciferase reporter assay was performed. This showed a significant decrease in luciferase activity when the Notch1 − 300 to − 250 region was deleted (Fig. 2c). To interrogate the Notch1 − 300 to − 250 region in more detail, the Notch1 − 300 to − 270 (N1 Δ − 300 to − 270) and − 280 to − 250 (N1 Δ − 280 to − 250) regions were deleted from the longest vector construct (N1 − 1140), and Jurkat T cells were transfected with each vector and stimulated with PMA/ionomycin. The N1 Δ − 300 to − 270-transfected cells exhibited a significant decrease in luciferase activity (Fig. 2d), whereas deletion of the Notch1 − 280 to − 250 region did not substantially affect the luciferase activity (Fig. 2d). To evaluate the importance of the identified cis element for TCR/CD28-mediated activation, Jurkat T cells were transfected with N1 − 1140 or N1 Δ − 300 to − 270 and stimulated using anti-CD3 and anti-CD28 antibodies. Deletion of the Notch1 − 300 to − 270 region considerably reduced the luciferase activity in cells stimulated using anti-CD3 and anti-CD28 antibodies (Fig. 2e). On the basis of these findings, we further interrogated the importance of the Notch1 − 300 to − 270 region in primary CD4+ T cells. The luciferase reporter vectors were transfected into mouse CD4+ T cells by electroporation, and the cells were cocultured with antigen-presenting cells in the presence of soluble anti-CD3 and anti-CD28 antibodies. The luciferase activity of the cells was much lower when they were transfected with N1 − 250 or N1 Δ − 300 to − 270 (Fig. 2f). These findings suggest that the Notch1 − 300 to − 270 region is important for the transcription of Notch1 in primary mouse CD4+ T cells, as well as in Jurkat cells.
Identification of the key regulatory region of the putative Notch1 promoter. (a) Conservation map of the mouse Notch1 promoter region. (b) Schematic representation of the serially deleted Notch1 promoter vector constructs. (c–e) Luciferase reporter assays performed in activated Jurkat T cells for identification of the key regulatory region using (c) transfection with truncated Notch1 promoter reporter vectors, (d) transfection with internally deleted Notch1 promoter reporter vectors, and (e) activation with anti-CD3 and anti-CD28 antibodies. (f) Luciferase reporter assay performed in activated CD4+ T cells transfected with Notch1 promoter reporter vectors selected during steps c–e. Data are mean ± SD. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 (Student’s t-test).
NF-κB directly binds to the Notch1 − 300 to − 270 region
To identify the transcriptional regulators that directly interact with the identified promoter region, in silico analyses of the Notch1 − 300 to − 270 sequence were conducted using MatInspector and Transfac (Table 1 and Supplementary table 1). Transfac predicted SP1 to be the most likely molecule to bind to this region (Supplementary table 1). However, co-transfection of the SP1 expression vector with N1 − 1140 or N1 Δ − 300 to − 270 did not alter luciferase activity significantly (Supplementary Fig. 2). The analysis using MatInspector predicted that the NF-κB subunits p50 and RelA would be binding partners (Table 1). Interestingly, overexpression of p50 and RelA increased the transcriptional activity induced by the N1 − 1140 construct, but deletion of the Notch1 − 300 to − 270 region abolished this transcriptional activation (Fig. 3a). An electrophoretic mobility shift assay (EMSA) was then conducted to confirm that these RelA-containing NF-κB subunits bind to the Notch1 − 300 to − 270 sequence via specific sequences. Nuclear extracts of p50- and RelA-overexpressing HEK293T cells interacted with the Notch1 − 300 to − 270 sequence probe (Fig. 3b), and addition of an anti-RelA antibody disrupted formation of the probe-NF-κB complex (Fig. 3b). In addition, we assessed binding of the RelA-containing NF-κB subunits to the Notch1 − 300 to − 270 sequence in the Jurkat T cell system, and found that nuclear extracts from activated Jurkat T cells bound to the Notch1 − 300 to − 270 sequence probe; in addition, formation of the probe-NF-κB complex was disrupted by an anti-RelA antibody (Fig. 3c). These data imply that sequence-specific direct binding of RelA-containing NF-κB proteins to the Notch1 − 300 to − 270 region occurs in activated T cells.
Validation of the predicted binding proteins for the Notch1 − 300 to − 270 sequence. (a) Luciferase reporter assay performed in activated Jurkat T cells that were overexpressing RelA or p50. (b, c) An electrophoretic mobility shift assay was performed using a Notch1 − 300 to − 270 sequence probe, and the probe-NF-κB complex was disrupted by an anti-RelA antibody in (b) RelA- and p50-overexpressing HEK293T cells, and (c) in activated Jurkat T cells. The original blots are presented in Supplementary Fig. 2. Data are presented as the mean ± SD. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 (Student’s t-test). N.S., not significant.
NF-κB upregulates Notch1 transcription in activated T cells
The physiological relevance of the interaction of NF-κB with the Notch1 − 300 to − 270 region was next evaluated using a chromatin immunoprecipitation (ChIP) assay. Crosslinked and sheared chromatin fragments obtained from activated Jurkat T cells were precipitated using anti-RelA antibody, and then quantitative real-time PCR analysis was used to assess the activity of the Notch1 − 300 to − 270 region using specific primers (Fig. 4a). This showed that NF-κB binds to the chromatin structure of the Notch1 − 300 to − 270 region in activated T cells, and this binding occurred in both living cells and under molecular experimental conditions. Moreover, overexpression of RelA and p50 increased Notch1 mRNA expression in Jurkat T cells (Fig. 4b); in addition, surface expression of Notch1 was increased by overexpression of these proteins (Fig. 4c, d). To further investigate the underlying mechanism(s), we exposed primary CD4+ T cells to SN50, an inhibitor of NF-κB activation, during CD3/CD28-mediated activation. Surface expression of Notch1 by vehicle-treated cells increased markedly after activation; however, this was abolished by treatment with SN50 (Fig. 4e, f). In addition, SN50 significantly decreased Notch1 transcription in activated primary CD4+ T cells (Fig. 4g). Furthermore, a ChIP assay using activated primary CD4+ T cells revealed that chromatin fragments containing the Notch1 − 300 to − 270 region were precipitated by an anti-RelA antibody, suggesting that Notch1 expression in activated primary CD4+ T cells is regulated by the same mechanism (Fig. 4h). Taken together, these observations suggest that NF-κB directly binds to the Notch1 − 300 to − 270 region and positively regulates the transcription of Notch1, leading to higher surface expression of Notch1.
Upregulation of Notch1 transcription by NF-κB in activated T cells. (a) Jurkat T cells activated with PMA/ionomycin were subjected to ChIP-qPCR analysis to detect chromatin containing the Notch1 -300–-270 region. N.D., not determined. (b) Quantitative real-time PCR for Notch1 mRNA expression in RelA and p50-overexpressing Jurkat T cells. (c, d) Flow cytometry for surface Notch1 expression in activated Jurkat T cells overexpressing RelA or p50. (e, f) Flow cytometry analysis of primary CD4+ T cells activated in the presence of 100 μM SN50 or PBS for the indicated times. (g) Quantitative real-time PCR of Notch1 mRNA from activated primary CD4+ T cells cultured for 48 h in the presence of 100 μM SN50 or PBS. (h) ChIP-qPCR analysis of primary CD4+ T cells, activated with anti-CD3 and anti-CD28 antibodies for the indicated times, to detect chromatin containing the Notch1 − 300 to − 270 region. Data are presented as the mean ± SD. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 (Student’s t-test).
Discussion
The importance of Notch signaling during T cell development has been well defined. Critically, deletion or inactivation of Notch1 causes development of the B cell lineage, rather than the T cell lineage, in mouse model systems11,12. In addition, Notch1 is upregulated in CD4+ T cells upon stimulation using anti-CD3 and anti-CD28 antibodies22. We have shown that the surface expression and mRNA expression of Notch1 are highly dependent on the CD28-PI3K pathway in activated T cells. PDK1 is regarded as the principal downstream effector of PI3K in CD4+ T cells, which implies that Notch1 plays a role in the PI3K-PDK1 downstream signaling complex and has roles in CD4+ T cell activation via PI3K-PDK1 downstream signaling cascades. Several studies of the roles of Notch1 in mature T cells have shown that Notch1 positively regulates expression of cytokines, interferon-γ, and interleukin-1022,23. However, conflicting data have been obtained regarding the roles of Notch1 during CD4+ T cell activation and proliferation. Some previous studies of the roles of Notch1 in CD4+ T cell activation and proliferation show stimulatory effects23,24,25, whereas others show inhibitory effects9,26. Therefore, a more detailed assessment regarding the exact roles of Notch1 in mature CD4+ T cell activation is required.
NF-κB participates in multiple processes in CD4+ T cells, including in their development, activation, survival, and differentiation27. The present findings suggest that NF-κB binds directly to the Notch1 promoter and upregulates its transcription during cellular activation. This is important because stimulation of the TCR/CD28 complex is fundamental for most T cell activities, and because NF-κB is one of the principal downstream effectors of the CD28-PI3K-PDK1 signaling pathway28,29. For example, CD4-specific deletion of Pdk1 impairs IκBα degradation under anti-CD3 and anti-CD28 simulation conditions29, and a recent study shows that PDK1-mediated NF-κB activation is essential for maintenance of regulatory T cells30. We showed that the inhibition of NF-κB activation by SN50 robustly disrupts Notch1 transcription during primary CD4+ T cell activation, although issues such as the possible effects of SN50-mediated inhibition of AP-1 and NFAT31 on Notch1 expression remain unaddressed. Furthermore, we confirmed binding of NF-κB to the Notch1 promoter region. In addition, Notch1 regulates NF-κB activity. Inactivation of Notch1 inhibits sustained NF-κB activity in T cells, and nuclear localization of NF-κB is regulated by the Notch1 intracellular domain32,33. Therefore, the interaction between Notch1 and NF-κB may represent a feedback mechanism whereby the activity or expression of each molecule is maintained, and might drive T cell activation or other downstream processes. However, it is unclear whether such feedback regulation would have positive or negative effects on T cell activities, because the roles of Notch1 in CD4+ T cell activation and proliferation are as yet poorly defined. Furthermore, the results of another study suggest an opposing effect of Notch1 on NF-κB activity34.
Activating mutations of Notch1 are regarded as a key diagnostic target for T-ALL, because uncontrolled activation of Notch1 signaling could be oncogenic35. In addition, the constitutive activation of NF-κB in T-ALL cells36 and higher expression of Notch1 in patients with T-ALL37 have been reported. As discussed above and as indicated by the present and previous findings, feedback regulation between Notch1 and NF-κB may occur, including in T-ALL cells. Interruption of this feedback regulation through targeting of the interaction of Notch1 with NF-κB may therefore represent a means of reducing Notch1 expression. The therapies for T-ALL have improved in recent years, and these include several inhibitors that target molecules that are essential for Notch1 signaling, such as γ‑secretase inhibitors, ADAM inhibitors, SERCA inhibitors, and monoclonal antibodies that target Notch138. However, their use might be associated with problems, including toxicity and limited efficacy, and a complete cure is difficult to achieve in some patients because of relapse or resistance to the therapy. Therefore, more diverse approaches to the regulation of Notch1 activity should be developed, such as transcriptional or epigenetic approaches. For example, modulation of the transcription of Notch1 might have synergistic effects with those of other drugs. However, most previous studies regarding Notch1 transcriptional regulation did not use a T cell system, and suggested a broad range for the Notch1 promoter sequence. Therefore, a deeper understanding of the regulatory mechanisms in T cells may provide clues regarding alternative therapeutic approaches for T-ALL.
We have shown that the expression of Notch1 is greatly increased during T cell activation, and that the Notch1 − 300 to − 270 region is critical for the positive regulation of Notch1 transcription in activated T cells. Moreover, NF-κB proteins, which are generally regarded as being downstream of PDK1, have been identified as transcriptional activators that directly bind to the Notch1 − 300 to − 270 region. Thus, upregulation of Notch1 is mediated via the PI3K-PDK1-NF-κB signaling pathway in activated T cells. Furthermore, the present findings were not made solely in a lymphoblast cell line: similar findings were made in primary CD4+ T cells isolated from mice. A better understanding of the detailed mechanisms of the transcriptional regulation of Notch1 in T cells may provide opportunities for further progress in research into autoimmune diseases and immunomodulatory therapies.
Methods
Vectors and mice
The Notch1 start codon (+ 1) to − 1140 region was amplified from mouse genomic DNA by PCR and subcloned into the pGL3 basic (N1 − 1140 vector). The N1 − 800, − 600, − 400, − 350, − 300, − 250, − 200, and − 150 vectors were constructed by truncating the N1 − 1140 vector by PCR and subcloning into the pGL3 basic vector. The forward primers used for constructing the vectors were as follows: N1 − 1140: 5′-GGGACGCGTGAGATAAAGCATGAGAGGCT-3′; N1 − 800: 5′-TTCACGCGTAGCCCTAGGGACAGG-3′; N1 − 600: 5′-TCCACGCGTATGTCGCAGAGGGCG-3′; N1 − 400, 5′-TTCACGCGTCCACCAAAGCGCTGG-3′; N1 − 350, 5′-TTCACGGCTGGAACCAGGGGCGGA-3′; N1 − 300, 5′-TTCACGCGTCATAGGGGCGGAG-3′; N1 − 250, 5′-TTCACGCGTCGGGAGGGAGCGCA-3′; N1 − 200 5′-TTCACGCGTGTGCGAGCGCAGTGA-3′; N1 − 150, 5′-TTCACGCGTCGCTGAGAGCCCAGC-3′; and 5′-TTGAAGCTTGCCTGCTCGCCAGCTGCC-3′ was used as the common reverse primer. The N1 Δ − 300 to − 270 and N1 Δ − 280 to − 250 vectors were constructed by deletion of these regions from the N1 − 1140 vector. pRenilla, pcDNA3-p50, pCMV-RelA, and pMigR1 vectors were used for transfection.
Pdk1flox/flox Cd4-Cre mice were generated as previously described29. The mice were held in a specific pathogen-free facility at Gwangju Institute of Science and Technology. Eight-to-ten-week-old wild-type C57BL/6 mice, Pdk1flox/flox Cd4-Cre, and Pdk1+/flox Cd4-Cre mice were euthanized by CO2 inhalation prior to use in the experiments. All animal experiments were approved by the Seoul National University’s Institutional Animal Care and Use Committee (SNU-210315-7-5) and were performed in accordance with relevant guidelines and regulations. The study was carried out in compliance with the ARRIVE guidelines.
Cell culture and flow cytometry
HEK293T cells were cultured in DMEM supplemented with 10% FBS and Jurkat cells, and mouse CD4+ T cells were cultured in RPMI-1640 supplemented with 10% FBS. Jurkat cells were activated by adding 500 ng/ml PMA and 100 ng/ml ionomycin to the culture medium, or anti-human CD3 and anti-human CD28 antibodies. Mouse CD4+ T cells were isolated from the spleens and lymph nodes of C57BL/6 mice using a negative isolation kit (Stem Cell Technologies, Vancouver, Canada). To activate the mouse CD4+ T cells, they were cultured in plates to which anti-mouse CD3 and anti-mouse CD28 antibodies were bound. Inhibitors were used at the following concentrations: 50 μM for LY294002 and 100 μM for SN50. Antigen-presenting cells were sorted from mouse mononuclear cells and then cocultured with mouse CD4+ T cells. Cell lines were transfected with FuGENE HD (Promega, Madison, WI, USA) or Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s protocol. The electroporation of mouse CD4+ T cells was performed using a Nucleofector kit for mouse T cells (Lonza, Basel, Switzerland).
Jurkat cells were co-transfected with pMigR1 to select transfected cells, which were sorted according to their GFP expression. Mouse cells were mixed with monoclonal antibodies purchased from eBioscience: CD3ε-PerCP/Cy5.5, CD4-eFlour 450, TCRβ-FITC, and Notch1-APC. These antibodies were used at a 1:200 dilution in PBS containing 2% serum. The cells were acquired using a FACSAria III sorter (BD, Franklin Lakes, NJ, USA) or a Guava EasyCyte analyzer (Merck Millipore), and the data were analyzed using FlowJo software (BD).
Nuclear fractionation
Cells were harvested and collected in tubes, and the cell pellets were washed, resuspended in a hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, and 1 μg/ml leupeptin), and then incubated on ice for 15 min. NP-40 was added to 0.05%, and the mixture was vigorously vortexed. The supernatant was removed after centrifugation at 1000 × g for 5 min at 4 °C, and the remaining pellet was washed with hypotonic buffer, resuspended in a hypertonic buffer (20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 1 μg/ml leupeptin), and incubated on ice for 30 min, with vigorous vortexing. Nuclear extracts were obtained by centrifugation at 20,000 × g for 10 min at 4 °C.
In silico analysis
Conserved sections of the Notch1 promoter region were searched for using Genome Browser (https://genome.ucsc.edu/). MatInspector (https://www.genomatix.de/matinspector/) and Transfac (https://genexplain.com/transfac/) were used to identify transcription factors that might bind to the Notch1 − 300 to − 270 region.
Quantitative real-time PCR
RNA was isolated from Jurkat or mouse CD4+ T cells using a RNeasy mini kit (Qiagen, Hilden, Germany). Up to 1 μg of RNA was converted to cDNA by reverse transcription (Enzynomics, Daejeon, Korea). SYBR green 2 × premix (Enzynomics) was used for quantitative real-time PCR on a CFX Connect real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA). Ct values were calculated for each target gene using a standard curve, and expression is stated relative to that of Gapdh.
Dual luciferase reporter assay
Ten thousand cells per well were seeded onto a 24-well plate and transfected with a reporter plasmid mixture (composed of pGL3 basic-based Notch1 vector and pRenilla) and other overexpression vectors. The cells were activated using PMA/ionomycin or anti-CD3 and anti-CD28 antibodies. They were harvested by centrifugation at 500 × g for 5 min at 4 °C and then washed twice using PBS. The luciferase activity of the samples was measured using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
Electrophoretic mobility shift assay
Nuclear extracts of NF-κB-transfected HEK293Ts and activated Jurkat cells were prepared as described above. The Notch1 − 300 to − 270 sequence was synthesized as a biotinylated probe (Macrogen, Seoul, Korea), and then the nuclear extracts and biotinylated probes were incubated together at room temperature. An anti-RelA antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was added to the mixture to disrupt formation of the probe-NF-κB complex. The mixtures were then separated on a 6% native polyacrylamide gel and transferred to a nylon membrane, which was incubated at 80 °C to permit crosslinking. The biotinylated DNA probes were then detected using a LightShift® Chemiluminescent EMSA Kit (Thermo Fisher Scientific) and exposure to X-ray films.
Chromatin immunoprecipitation assay
Crosslinked chromatin was extracted from 2 × 107 Jurkat cells or 5 × 106 mouse primary CD4+ T cells and sonicated to shear the DNA, and then sheared crosslinked chromatin DNA was incubated with protein G agarose slurry for 1 h at 4 °C, with gentle rotation. After removing the agarose by centrifugation, the precleared supernatant was incubated with 5 μg anti-RelA antibody overnight at 4 °C, with gentle rotation. Positive control samples were incubated with 1 μg of anti-RNA polymerase II antibody or anti-histone H3 antibody, and negative control samples were incubated with 1 μg of normal mouse IgG. Protein G agarose was then added to each sample, which was incubated for 1 h at 4 °C with gentle rotation. Protein G agarose was washed and put through elution. In addition, the purified DNA was used for quantitative real-time PCR analysis. ChIP was performed using an EZ-ChIP kit (Merck, Darmstadt, Germany), according to the manufacturer’s protocol.
Statistical analysis
The significance of differences between two groups was analysed using Student’s t-test. All data are expressed as the mean ± SD. For all statistical tests, P-values ≤ 0.05 were considered significant.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
MacDonald, H. R., Wilson, A. & Radtke, F. Notch1 and T-cell development: insights from conditional knockout mice. Trends Immunol. 22, 155–160. https://doi.org/10.1016/s1471-4906(00)01828-7 (2001).
Blaumueller, C. M., Qi, H., Zagouras, P. & Artavanis-Tsakonas, S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90, 281–291. https://doi.org/10.1016/s0092-8674(00)80336-0 (1997).
Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M. E. Notch signaling. Science 268, 225–232. https://doi.org/10.1126/science.7716513 (1995).
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776. https://doi.org/10.1126/science.284.5415.770 (1999).
Shutter, J. R. et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 14, 1313–1318 (2000).
De Strooper, B. et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522. https://doi.org/10.1038/19083 (1999).
Mumm, J. S. et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206. https://doi.org/10.1016/s1097-2765(00)80416-5 (2000).
van Tetering, G. et al. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284, 31018–31027. https://doi.org/10.1074/jbc.M109.006775 (2009).
Eagar, T. N. et al. Notch 1 signaling regulates peripheral T cell activation. Immunity 20, 407–415. https://doi.org/10.1016/s1074-7613(04)00081-0 (2004).
Ghaedi, M. et al. Common-lymphoid-progenitor-independent pathways of innate and T lymphocyte development. Cell Rep 15, 471–480. https://doi.org/10.1016/j.celrep.2016.03.039 (2016).
Pui, J. C. et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308. https://doi.org/10.1016/s1074-7613(00)80105-3 (1999).
Radtke, F. et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10, 547–558. https://doi.org/10.1016/s1074-7613(00)80054-0 (1999).
Tanigaki, K. et al. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20, 611–622. https://doi.org/10.1016/s1074-7613(04)00109-8 (2004).
Washburn, T. et al. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell 88, 833–843. https://doi.org/10.1016/s0092-8674(00)81929-7 (1997).
Van de Walle, I. et al. An early decrease in Notch activation is required for human TCR-alphabeta lineage differentiation at the expense of TCR-gammadelta T cells. Blood 113, 2988–2998. https://doi.org/10.1182/blood-2008-06-164871 (2009).
Belver, L. & Ferrando, A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat. Rev. Cancer 16, 494–507. https://doi.org/10.1038/nrc.2016.63 (2016).
Lefort, K. et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 21, 562–577. https://doi.org/10.1101/gad.1484707 (2007).
Yugawa, T. et al. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 27, 3732–3742. https://doi.org/10.1128/MCB.02119-06 (2007).
Lambertini, C., Pantano, S. & Dotto, G. P. Differential control of Notch1 gene transcription by Klf4 and Sp3 transcription factors in normal versus cancer-derived keratinocytes. PLoS One 5, e10369. https://doi.org/10.1371/journal.pone.0010369 (2010).
Jia, B. et al. SIRT6 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells through antagonizing DNMT1. Front. Cell Dev. Biol. 9, 648627. https://doi.org/10.3389/fcell.2021.648627 (2021).
Zhang, X. et al. DLX5 promotes osteosarcoma progression via activation of the NOTCH signaling pathway. Am. J. Cancer Res. 11, 3354–3374 (2021).
Benson, R. A. et al. Notch1 co-localizes with CD4 on activated T cells and Notch signaling is required for IL-10 production. Eur. J. Immunol. 35, 859–869. https://doi.org/10.1002/eji.200425562 (2005).
Palaga, T., Miele, L., Golde, T. E. & Osborne, B. A. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J. Immunol. 171, 3019–3024. https://doi.org/10.4049/jimmunol.171.6.3019 (2003).
Adler, S. H. et al. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 171, 2896–2903. https://doi.org/10.4049/jimmunol.171.6.2896 (2003).
Joshi, I. et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 113, 1689–1698. https://doi.org/10.1182/blood-2008-03-147967 (2009).
Rutz, S., Mordmuller, B., Sakano, S. & Scheffold, A. Notch ligands Delta-like1, Delta-like4 and Jagged1 differentially regulate activation of peripheral T helper cells. Eur. J. Immunol. 35, 2443–2451. https://doi.org/10.1002/eji.200526294 (2005).
Oh, H. & Ghosh, S. NF-kappaB: roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 252, 41–51. https://doi.org/10.1111/imr.12033 (2013).
Hwang, J. R., Byeon, Y., Kim, D. & Park, S. G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 52, 750–761. https://doi.org/10.1038/s12276-020-0435-8 (2020).
Park, S. G. et al. The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-kappaB and activate T cells. Nat. Immunol. 10, 158–166. https://doi.org/10.1038/ni.1687 (2009).
Oh, H. et al. PDK1 is required for maintenance of CD4(+) Foxp3(+) regulatory T cell function. J. Immunol. 206, 1776–1783. https://doi.org/10.4049/jimmunol.2000051 (2021).
Torgerson, T. R., Colosia, A. D., Donahue, J. P., Lin, Y. Z. & Hawiger, J. Regulation of NF-kappa B, AP-1, NFAT, and STAT1 nuclear import in T lymphocytes by noninvasive delivery of peptide carrying the nuclear localization sequence of NF-kappa B p50. J. Immunol. 161, 6084–6092 (1998).
Shin, H. M. et al. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 25, 129–138. https://doi.org/10.1038/sj.emboj.7600902 (2006).
Espinosa, L. et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell 18, 268–281. https://doi.org/10.1016/j.ccr.2010.08.006 (2010).
Wang, J. et al. Human Notch-1 inhibits NF-kappa B activity in the nucleus through a direct interaction involving a novel domain. J. Immunol. 167, 289–295. https://doi.org/10.4049/jimmunol.167.1.289 (2001).
Ferrando, A. A. The role of NOTCH1 signaling in T-ALL. Hematol. Am. Soc. Hematol. Educ. Program 2009, 353–361. https://doi.org/10.1182/asheducation-2009.1.353 (2009).
Kordes, U., Krappmann, D., Heissmeyer, V., Ludwig, W. D. & Scheidereit, C. Transcription factor NF-kappaB is constitutively activated in acute lymphoblastic leukemia cells. Leukemia 14, 399–402. https://doi.org/10.1038/sj.leu.2401705 (2000).
Lin, C. et al. Mutations increased overexpression of Notch1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 12, 13. https://doi.org/10.1186/1475-2867-12-13 (2012).
Cordo, V., van der Zwet, J. C. G., Cante-Barrett, K., Pieters, R. & Meijerink, J. P. P. T-cell acute lymphoblastic Leukemia: a roadmap to targeted therapies. Blood Cancer Discov. 2, 19–31. https://doi.org/10.1158/2643-3230.BCD-20-0093 (2021).
Acknowledgements
This research was supported by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI22C0828), and by the National Research Foundation of Korea (Grant Numbers: NRF-2021R1A2C3011211 and NRF-2022M3A9I2017587).
Author information
Authors and Affiliations
Contributions
Conceptualization: D.K., S.G.P.; data curation: J.R.H., D.K., J.A.K., S.H.P.; formal analysis: J.R.H., S.G.P.; writing - original draft: J.R.H.; writing - review and editing: S.G.P. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hwang, JR., Kim, D., Kang, JA. et al. Transcriptional regulation of Notch1 by nuclear factor-κB during T cell activation. Sci Rep 13, 43 (2023). https://doi.org/10.1038/s41598-022-26674-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26674-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.