Abstract
The genus Chelus, commonly known as Matamata is one of the most emblematic and remarkable species among the Neotropical chelids. It is an Amazonian species with an extensive distribution throughout Negro/Orinoco and Amazonas River basins. Currently, two species are formally recognized: Chelus orinocensis and Chelus fimbriata and although it is still classified as "Least Concern" in the IUCN, the Matamatas are very appreciated and illegally sold in the international pet trade. Regardless, little is known regarding many aspects of its natural history. Chromosomal features for Chelus, for instance, are meagre and practically restricted to the description of the diploid number (2n = 50) for Chelus fimbriata, and its sex determining strategies are yet to be fully investigated. Here, we examined the karyotype of Chelus fimbriata and the newly described Chelus orinocensis, applying an extensive conventional and molecular cytogenetic approach. This allowed us to identify a genetic sex determining mechanism with a micro XY sex chromosome system in both species, a system that was likely present in their most common recent ancestor Chelus colombiana. Furthermore, the XY system found in Chelus orinocensis and Chelus fimbriata, as seen in other chelid species, recruited several repeat motifs, possibly prior to the split of South America and Australasian lineages, indicating that such system indeed dates back to the earliest lineages of Chelid species.
Similar content being viewed by others
Introduction
The side-necked turtles from the Chelidae family represent one of the three main living lineages that make up the Pleurodira suborder. They have origin in South America, with fossil records dating from Gondwanic events1,2,3. Several studies have shown that chelid turtles diversified even before the split of Gondwana4,5,6,7, suggesting that besides vicariant events, dispersal events also drove the diversification of these species in the beginning of the fragmentation of Gondwana3. Such events explain the current distribution of the approximately 60 extant species of Chelidae that are restricted to the Southern Hemisphere, occurring throughout Australasia and South America8,9,10,11.
The Amazon region is considered a hotspot and primary source of Neotropical biodiversity12, including for species of freshwater turtles13,14, with groups that are widely distributed along the Amazon hydrographic basins, such as turtles of the genera Chelus and Mesoclemmys15,16.
The genus Chelus, or simply Matamata, is clearly one of the most emblematic and charismatic species among the Neotropical chelids, being considered one of the most bizarre species in the world due to its particular lifestyle and unusual external morphology, such as triangular head, tiny eyes, extremely elongated neck and a long tubular nose. It is also among the largest species of the Chelidae family17,18. Besides, Matamatas are one of the few species in the world that are predominantly carnivorous, with a diet almost exclusively based on live fish19,20.
The Matamata, for centuries was considered as a monotypic species with a wide distribution, however, recently genetic and morphological analyzes revealed the existence of at least two different species occurring in different waterscapes along its extensive distribution in the Amazon region, namely: Chelus orinocensis and Chelus fimbriata21. C. orinocensis occurs primarily along the upper Negro and Orinoco River Basins, while C. fimbriata occurs in Solimões/Amazonas River Basin16 and both species date back to the late Miocene, with recent 13my of independent divergence21.
Despite the Matamatas are one of the most well-known species to the general public, especially due to its peculiar morphology and appreciation in the international pet trade8,22, many aspects of their natural history still remain unknown. Chromosomal features, for instance, are scant and practically limited to the description of the diploid number (2n = 50) for Chelus fimbriata23,24,25. Chelids stands out as the most diverse group among Pleurodira, exhibiting several karyotype configurations, varying including in number of macrochromosomes (Mac) and microchromosomes (mic), as well as the presence of differentiated and undifferentiated sex chromosomes24,26,27,28. Most species have the GSD (genetic sex determination) mechanism as mode of sex determination29,30, however, despite the evidence of one bivalent with no pairing in meiosis of Chelus fimbriata23, the sex determining strategies of Matamatas remain unanswered, as well as whether the species have sex chromosome systems, as observed in the vast majority of species in the family27,28,29,31,32,33. In this sense, a more detailed investigation into the karyotype composition of the two species of Chelus is required, especially to investigate the sex determination strategies and presence of Mac or mic sex chromosomes of this eccentric amazonian turtle species.
In this study, we aimed to provide a full description of the karyotype composition for Chelus orinocensis and Chelus fimbriata from the Amazon Rainforest. For that, we applied multiple conventional (Giemsa staining and C-banding) and molecular (Comparative Genomic Hybridization (CGH), FISH mapping of telomeric (TTAAGG)n, 18S rDNA and Simple Short Repeats (SSRs) sequences) cytogenetic tools. Also, we identified sex chromosomes and modes of sex determination in these charismatic turtles and discussed the evolution of sex chromosomes and the role of repetitive sequences along their diversification in the Amazon region.
Results
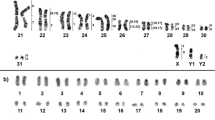
Karyotype and C-positive heterochromatin
The Matamatas have 2n = 50 chromosomes with 22 Mac and 28 mic. The karyotype is composed by 6m + 4sm + 4st + 8a and 28mi (mic predominantly acrocentrics) for both species, with a fundamental number (NF) equal to 64 for males and females (Fig. 1). In Chelus fimbriata, the C-positive heterochromatin was found in few chromosome pairs (4, 6, 7, 8, 9, 10 and 12) (Fig. 1A), with a preferential accumulation on the centromeric regions, excepting the 10th pair, which had the short arm completely heterochromatic. In contrast, Chelus orinocensis exhibited a greater amount of C-positive heterochromatin (pairs 1–6, 8, 9, 10, 12, 13, 14, 16 and 19) (Fig. 1B), with conspicuous markings predominantly on centromeric position, but also on telomeric regions (pair 1) and on terminal position of the 5th chromosome pair. The mic pairs (12, 13, 14, 16 and 19) had only centromeric markings.
Mapping of SSRs motifs, 18S rDNA and Telomeric repeats
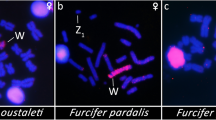
All eighteen microsatellite repeat motifs used (AC)15, (AT)15, (GT)15, (AG)15, (AGC)10, (AAT)10, (CGG)10, (AAC)10, (GATA)8, (GACA)8, (ACGC)8, (GGAT)8, (AATG)8, (AAGG)8, (ATCC)8, (AATC)8, (AAAC)8 and (AAAAT)8 showed hybridization signals on chromosomes of Chelus orinocensis and Chelus fimbriata (Figs. 2, 3, 4, 5, 6, 7). We found the same pattern of SSRs amplification for both species, including male-specific pattern of accumulation for some SSRs, which identifies Chelus orinocensis and Chelus fimbriata as a GSD species with a mic XY sex chromosome system and with X and Y-linked repeats (Figs. 2, 3, 4, 5, 6, 7). The (AC)15, (AG)15, (GATA)8, (AAGG)8 and (AAAAT)8 showed hybridization signals on both Mac and mic of males and females, but with a particular amplification on the Y sex chromosome, excepting for the (GATA)8, that was found to show a unique pattern (Figs. 3, 6), three markings in males (the 9th pair and a single mic) and four markings in females (the 9th pair and two mics), this reveals a particular amplification in females and identifies the X chromosome. On the other hand, the motifs (AT)15, (GT)15, (AGC)10, (AAT)10, (CGG)10, (AAC)10, (GACA)8, (ACGC)8, (GGAT)8, (AATG)8, (ATCC)8, (AATC)8 and (AAAC)8 were found to be male-specific, with exclusive amplification (Figs. 2, 3, 4, 5, 6, 7). Besides, we were able to properly identify the X chromosome with specific accumulation of (GATA)8, even with the morphological similarity of the X with many other mics. Interestingly, even with particular accumulation of several SSR motifs, the Y chromosomes of Chelus orinocensis and Chelus fimbriata are not heterochromatic (Fig. 1), being identified solely through the mapping of the SSRs.
The mapping of the 18S rDNA sequences showed simple markings on the secondary constriction of the second largest acrocentrics of the complement, the 8th pair in both species (Fig. 8). The telomeric motifs was evidenced on all terminal portions of all Mac and mic, with no evidence of ITSs (i.e., tandemly repeats of TTAGGGn sequence that are localized at intrachromosomal portions such as centromeres and interstitial positions and referred as Insterstitial Telomeric Sequences), nonetheless, a clear amplification of such sequences was evidenced on XY chromosomes (Fig. 8). The (TTAGGG)n amplification on X and Y was confirmed by the subsequently mapping of (GATA)n and (GACA)n respectively, in the same metaphase spread in males and females (picture not shown). Because the hybridization patterns of 18S rDNA and telomeric repeats for males and females were exactly the same, we selected a male representative metaphase to illustrate the results.
Comparative genomic hybridization (CGH)
Our intraspecific comparison between males and females of Chelus orinocensis and Chelus fimbriata produced intense hybridization signals on centromeric position of some Mac, not necessarily collocated with heterochromatic portions (Fig. 9). Some mic showed scattered markings in both species, however, the merged images identified male-specific sequences accumulated in a tiny mic of the karyotype, the micro Y sex chromosome (Fig. 9). Since the hybridization signals on female metaphase spreads showed only shared sequences, we selected only male’s metaphase of Chelus orinocensis and Chelus fimbriata to illustrate the results and highlight the Y-linked markings. Sequential detection of C-positive heterochromatin in Chelus orinocensis and Chelus fimbriata in fact revealed that the Y chromosome in both species is not heterochromatic (image not shown).
Discussion
In the present study, we cytogenetically analyzed the two extant species of Matamatas, the newly described Chelus orinocensis and reinvestigate the karyotypic composition of Chelus fimbriata since its first description almost half a century ago. We discuss hypotheses and trends on the evolution of sex chromosomes and the stability of the XY system that likely dates back to the origins of the Chelidae family ~ 103–160 Mya. Our findings corroborate previous mention in the literature regarding the karyotype composition of Chelus23,24. We also detected 2n = 50 chromosomes for the two species of Matamatas (Fig. 1), highlighting gross conservation of karyotypes between species, differing from each other only by the C-banding pattern (Fig. 1). Our study also provided evidence that both species have GSD with XY mic sex chromosomes.
In the late 70s, Barros and colleagues published the first karyotype description for the genus Chelus, (fairly similar to our findings), which had 2n = 50 chromosomes and no difference between males and females. However, the evidence of heterochromatic portions and one bivalent without pairing in meiosis already indicated that the species would be GSD with XY sex chromosomes system. Confirmation of the presence of sex chromosome in Chelus came just nearly half a century later with the findings from our study, where we evidenced a GSD mechanism as mode of sex determining and mic XY chromosomes in both analyzed species, similar to what has been described in Australian Chelid species.
Modes of sex determining and sex chromosomes exhibit a striking evolutionary dynamic in vertebrates, with complex mechanisms that have evolved independently and multiple times across lineages and tempo34,35,36,37,38. Several reptile lineages follow the mainstream with multiple turnovers even in closely related groups39,40. Turtles also accompany this evolutionary path, with transitions among mechanisms of sex determining (Environmental sex determining—ESD/Genetic sex determining—GSD) and systems of sex chromosomes (XY/ZW) that evolved repeatedly along its evolutionary history41,42,43,44 and several intra/interchromosomal rearrangements are signed as the main source of such diversity31,32,45,46,47,48. In general, the GSD mechanism appears to be the sex determination mode chosen by the Neotropical and Australasian chelids since split from their sister families Podocnemididae and Pelomedusidae (both ESD/Temperature-dependent sex determination) (Fig. 10)6,21,29,30,49.
The ancestral state of Mac or mic sex chromosomes in Australasian chelids still remains as an open question27,32,33,50, although the most probabilistic scenario encompasses translocation/fusion events from mic to Mac sex chromosomes in the ancestor of the clades with Mac sex chromosomes (species with 2n = 50) and the clade with mic sex chromosomes (Chelodina spp. with 2n = 54)32,33,48,50. In Neotropical lineages, out of the three species analyzed using more refined cytogenetic tools, all have mic X [28, present study]. The mic XY of the Red Side-Necked Turtle Rhinemys rufipes dates back the Oligocene28 and much likely share the same origin with the XY of Chelus orinocensis and Chelus fimbriata, that goes back to the late Cretaceous/Paleocene6,21. The unquestionable South American origins of Chelidae1,2,3, and the likely ancestry of such mic sex chromosome in Neotropical lineages suggest that it was already present since the genesis of Chelidae in the Upper Jurassic ~ 103–160 Mya, highlighting an ancient and long-term XY system along the evolutionary history of these turtles. However, an overview of sex chromosome trends in a broader of neotropical chelids is still required to fulfill into this gap regarding the ancestor state in the family as a whole, as well as to uncover whether Mac or mic XY sex chromosome is present in the former Chelidae lineages. Test the homology of the known mic sex chromosomes of neotropical lineages (Chelus and Rhinemys) with those Mac and mic sex chromosomes of Australasia lineages is an essential step to highlight such ancestry.
The Chelidae chromosomes vary in number (including quantity of Mac and mic) and morphology of chromosomes25,27,28 and much of this diversity, as already mentioned, is attributed to several inter-intra chromosomal rearrangements that occurred along its evolution32,48,50. For example, the smallest metacentric pairs present in Chelus orinocensis and Chelus fimbriata (pairs 7 and 10) are missing in most Neotropical species with known karyotype23,24,26,28. With few exceptions (e.g., Mesoclemmys gibba 2 = 60, Mesoclemmys raniceps 2n = 40 and Mesoclemmys sp. 2n = 42), most Neotropical species have 2n = 58, where chromosomal fission events could be behind this diversity. If we consider centric fissions in the pairs 4, 5, 7 and 10 of the Chelus complement (2n = 50), this would result in exactly the same 2n = 58 and karyotypic formula seen in Rhinemys rufipes, Mesoclemmys vanderhaegei, Mesoclemmys perplexa, Mesoclemmys tuberculata, Mesoclemmys dahli, Mesoclemmys hogei, Mesoclemmys nasuta, Phrynops geoffroanus and Hydromedusa tectifera [24,25,28,51,52, personal data]. However, Hydromedusa tectifera (2n = 58) arose in the Middle Jurassic ~ 100 mya6, much older than 2n = 50 found in Chelus (Fig. 10). This raises two interesting and different evolutionary trajectories for chromosomal diversification of Neotropical chelids: (1) much older fusion events from the split of Hydromedusa and Chelus and, subsequently, fissions in the bi-armed pairs (4, 5, 7, 10) leading back to the 2n = 58 found in the most other neotropical chelids or (2), where the 2n = 58 would also represent the ancestral chromosome state and the 2n = 50 would represent a unique apomorphy arising solely in Chelus through fusion events from its most common recent ancestor after its split in the Middle Jurassic and the 2n = 58 was simply retained in most other Chelidae lineages. In fact, the 2n = 58 chromosomes seem to be the ancestral state for Chelidae, since the 2n = 50 appeared only twice along its evolution (in Chelus spp. with mic XY and in the Australasian lineages with Mac XY), possibly independently. Chromosomal paintings using the likely bi-armed chromosomes involved in these fusion/fission events is required to test the full homology across 2n = 50 and 2n = 58 chromosomes (as well as other 2n configurations in the family) and will certainly shed light on this evolutionary puzzle.
Despite these evident events of past chromosomal rearrangements orchestrating the karyotype diversification in Chelidae, with few exceptions27,28,53, relics of intra/interstitial telomeric-like repeats are rarely detected in turtles as a whole54. In Chelus spp. for example, no traits of ITSs have been found, but a particular amplification of telomeric-like sequences on XY chromosomes.
Amplification of telomeric repeats on sex chromosomes (XY and ZW) has been reported in a range of reptile lineages50,55,56, being frequently suggested as having regulatory activities and involved in heterochromatinization process. Telomeric-like repeats is clearly associated with Mac and mic sex chromosomes in Australasian chelids, including in a hybrid between Emydura and Elseya27. Clemente et al.54 mapped telomeric sequences in a male of Mesoclemmys hogei (South American lineage) and found no evidence of ITSs, but a tiny mic chromosome clearly showed a great accumulation of telomeric-like repeats, suggesting that this species might have an XY system, however, further analyzes with more males and females as well as the mapping of Y-linked SSRs (like those ones we have shown to be sex-specific in Chelus and in Rhinemys28 is necessary to confirm Mesoclemmys hogei as having mic XY chromosomes. In Chelus orinocensis and Chelus fimbriata, the amplification of telomeric-like repeats on XY, as well as the presence of several other SSRs sex-linked repeats apparently follow an alternative evolutionary path, not necessarily correlated to heterochromatinization process, since the XY in both species, unlike what was observed in Rhinemys rufipes28, were found to be non-heterochromatic. Perhaps, the recalcitrant recruitment of such repeats on these sex chromosomes might be related to a role beyond the regulation of the component of satellite DNA on XY, but also acting in the sex determining and in the maintenance of the dynamics of XY system architecture across Chelidae lineages.
Another repetitive and fundamental sequence, sometimes encountered in vertebrate sex chromosomes are the highly conserved rDNAs, which have a unique57,58,59,60,61 evolutionary dynamic, with multiple and crucial roles, acting from the control of cell aging/maintenance of genome integrity to shaping sex chromosomes, roles and functions far beyond ribosomal synthesis (for details on rDNA functioning, see Symonová62). In turtles, the 18S rDNA is frequently recruited on both ZW and XY systems27,63,64 and in Australasian chelids, such association is evidenced in both lineages with mic XY (2n = 58) and Mac XY (2n = 50)27. In Neotropical species, hitherto, no evidence of rDNAs accumulation on sex chromosomes has been found [28, present study] in addition, only Mac pairs harbor the rDNA sites in Rhinemys rufipes (pair 3), Chelus orinocensis and Chelus fimbriata (pair 8), all species with mic sex chromosomes. Seemingly, the accumulation of rDNAs on sex chromosomes is exclusive to Australasian species. Regardless, rDNAs are thought to somewhat represent an evolutionary driver for sex chromosome evolution in groups with which they are associated60,65, including Chelidae, where they have likely undergone multiple translocation events due to its association with other repetitive sequences (e.g., TEs/transposable elements and SSRs/simple short repeats), resulting in different chromosomal pairs carrying the rDNA. Interestingly, only one SSR motif (AG) was found bearing the 18S rDNA sites in C. orinocensis and C. fimbriata (Figs. 2, 5), while at least three (GT, AG, AAC) has been detected in Rhinemys rufipes28, probably reflective of different landscape of SSRs in these species.
SSRs represent an important and dynamic portion of genomes, that can evolve rapidly and independently66,67,68,69, being often associated with regulatory functions in genome architecture, structural activities in DNA, chromatin organization, gene expression and a myriad of other important functions70,71,72. This dynamic behavior, together with its expansion and contraction in genomes and high mutation rates remarkably reflects in distinct SSR landscapes, even in closely related species69,73,74,75,76,77,78.
Although turtles are naturally SSR-poor68,69,79, the frequent recruitment of such sequences to sex chromosomes indicates that SSRs repeats may be inherently shaping the evolution of sex chromosomes in chelids [27,28,50, present study] . The ATCC repeat motif, for example, is found on the sex chromosomes of at least 5 species of Chelidae [28,32, present sudy]. In Matamatas, thirteen SSRs are exclusively amplified on the tiny mic Y chromosome (Figs. 2, 3, 4, 5, 6, 7). Interestingly, six of them, found exclusively on sex chromosomes of Chelus spp. (GT, AAC, GACA, GGAT, ACGC, ATCC), are found in autosomes and on the mic Y of Rhinemys rufipes [28, present study], suggesting that in fact, closely related species may exhibit completely divergent SSR landscapes. In contrast, Chelus orinocensis and Chelus fimbriata shows exactly the same pattern of SSRs (Figs. 2, 3, 4, 5, 6, 7), which may be explained by their recent split (13Mya) and past genetic introgression and gene flow promoted by the wide distribution that these species had during the conformation of ancient Amazonian waterscapes over these million years of evolution21. This similarity of SSRs between the two Matamata species also suggests that the mic XY system could already be present in Chelus colombiana, the most recent common ancestor that gave rise to the two living species80.
Our study is the first to offer an extensive and more complete chromosomal characterization of the two living species of Matamatas with a gap of almost half a century since their first karyotype description. Our data surely brings important novelties and shed light on the evolutionary history and adds new pieces to the puzzle of chromosomal evolution in Chelidae. We discovery modes of sex determination and a mic XY sex chromosome system, often associated with particular SSR motifs, highlighting ancestry among species that probably dates back to the origin of chelids in the Upper Jurassic ~ 103–160 Mya (Fig. 10). Despite the very similar SSR landscape between the two Matamatas species, such repeats likely drove the pathways that resulted in the different karyotype configurations and sex chromosomes in South America and Australasian lineages. This study is part of a series on cytogenetics and cytogenomics investigation in Amazonian reptiles and their hidden evolutionary diversity.
Materials and methods
Sampling, mitotic chromosomes preparation and C-banding
The turtles were sampled from natural populations throughout their distribution, Chelus orinocensis in Negro River and Chelus fimbriata in Amazonas River. The collects were performed under permission granted by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) number: 45275. We analyzed 5 males and 4 females of Chelus orinocensis and 3 males and 3 females of Chelus fimbriata. Chromosomal preparations were cultured from small blood samples at 29 ℃ according to Viana et al.25. The C-positive heterochromatin was detected following Sumner81.
All experiments were performed in accordance with relevant guidelines and regulations. We declare that all procedures and experimental protocols were approved and performed under the rules of the Ethics Committee of the National Institute of Amazonian Research (Permission number: 018/2017). The study was carried out in compliance with the ARRIVE guidelines.
Probes for chromosome hybridization
The 18S rDNA and Telomeric (TTAGGG)n probes were isolated following Gross et al.82 and Ijdo et al.83, respectively. Both probes were labeled with Aminoallyl-dUTP ATTO-488 (green) by Nick-translation means (Jena Bioscience, Jena, Germany). Simple Short Repeats (SSRs), (AC)15, (AT)15, (GT)15, (AG)15, (AGC)10, (AAT)10, (CGG)10, (AAC)10, (GATA)8, (GACA)8, (ACGC)8, (GGAT)8, (AATG)8, (AAGG)8, (ATCC)8, (AATC)8, (AAAC)8 and (AAAAT)8 were used directly labeled with Cy-3 during the synthesis.
Fluorescence in situ hybridization (FISH) for repetitive DNA mapping
FISH followed the procedures detailed in our previous works28,84,85. Briefly, the chromosomes were denatured in 70% formamide/2xSSC at 70 °C, spreads were dehydrated in ethanol (100%). Then, 20 µl of the hybridization mixture (100 ng of each probe, 50% deionized formamide and 10% dextran sulfate) was dropped on the slides and the hybridization was carried out for 24 h at 37 °C in a moist chamber. The chromosomes were counterstained with DAPI (1.2 µg/ml) and mounted in antifade solution (Vector, Burlingame, CA, USA).
Preparation of probes for comparative genomic hybridization (CGH)
The gDNAs of males and females of Matamatas (Chelus orinocensis and Chelus fimbriata) were extracted from small blood samples using the Wizard Genomic Purification Kit (Promega), following the manufacturer’s recommendations. Female-derived gDNA was labeled with Aminoallyl-dUTP ATTO-550 (red) and male’ gDNA with Aminoallyl-dUTP ATTO-488 (green) using Nick-translation labeling Kit (Jena Bioscience, Jena, Germany). The final hybridization mixture for each slide (20 μl) was composed of male- and female-derived gDNAs (500 ng each), 25 μg of male-derived Cot-1 DNA (i.e. the fraction of genomic DNA enriched for highly repetitive sequences), prepared following86 and the hybridization buffer containing 50% formamide, 2 × SSC, 10% SDS, 10% dextran sulfate and Denhardt´s buffer, pH 7.0. The probes were ethanol-precipitated and the dried pellets were resuspended in hybridization buffer, as above mentioned.
Comparative Genomic Hybridization (CGH)
The intraespecific CGH experiments were performed according to our previous studies28,38,85,87. The slides were incubated at 37° C in a dark humid chamber for three days. The chromosomes were counterstained with DAPI (1.2 µg/ml) and mounted in an antifade solution (Vector, Burlingame, CA, USA).
Microscopic analyses
At least 10 metaphase spreads for male and females were analyzed to confirm the karyotype structure and FISH results. Images were captured using an Olympus BX51 microscope (Olympus Corporation). Chromosomes were classified as macrochromosomes (Mac) and microchromosomes (mic) or as metacentric (m), submetacentric (sm), subtelocentric (st), and acrocentric (a), according to Levan et al.88.
Ethics statement
All experiments were performed in accordance with relevant guidelines and regulations. We declare that all procedures and experimental protocols were approved and performed under the rules of the Ethics Committee of the National Institute of Amazonian Research (Permission number: 018/2017). The study was carried out in compliance with the ARRIVE guidelines.
Data availability
All data generated and analyzed during this study are included in this published article. No datasets were generated during the current study. Additional information about this study is available from the corresponding author upon reasonable request.
References
Ferreira, G. S. & Langer, M. C. A pelomedusoid (Testudines, Pleurodira) plastron from the Lower Cretaceous of Alagoas, Brazil. Cretaceous Res. 46, 267–271 (2013).
Romano, P. S. R., Gallo, V., Ramos, R. R. C. & Antonioli, L. Atolchelys lepida, a new side-necked turtle from the Early Cretaceous of Brazil and the age of crown Pleurodira. Biol. Lett. 10, 1–10 (2014).
Ferreira, G. S., Bronzati, M., Langer, M. C. & Sterli, J. Phylogeny, biogeography and diversification patterns of side-necked turtles (Testudines: Pleurodira). R. Soc. Open Sci. 5, 1–17 (2018).
de la Fuente, M. S., Umazano, A. M., Sterli, J. & Carballido, J. L. New chelid turtles of the lower section of the Cerro Barcino formation (Aptian-Albian?), Patagonia, Argentina. Cretaceous Res. 32, 527–537 (2011).
Joyce, W. G., Parham, J. F., Lyson, T. R., Warnock, R. C. M. & Donoghue, P. C. J. A divergence dating analysis of turtles using fossil calibrations: An example of best practices. J. Paleontol. 87, 612–634 (2013).
Pereira, A. G., Sterli, J., Moreira, F. R. R. & Schrago, C. G. Multilocus phylogeny and statistical biogeography clarify the evolutionary history of major lineages of turtles. Mol. Phylogenet. Evol. 113, 59–66 (2017).
Shaffer, H. B., McCartney-Melstad, E., Near, T. J., Mount, G. G. & Spinks, P. Q. Phylogenomic analyses of 539 highly informative loci dates a fully resolved time tree for the major clades of living turtles (Testudines). Mol. Phylogenet. Evol. 115, 7–15 (2017).
Rueda-Almonacid, J. Vicente. Las tortugas y los cocodrilianos de los países andinos de trópico (Conservación Internacional, 2007).
Georges, A. & Thomson, S. Diversity of Australasian freshwater turtles, with an annotated synonymy and keys to species. Zootaxa 2496, 1–37 (2010).
TTWG. Turtles of the World: Annotated Checklist and Atlas of Taxonomy, Synonymy, Distribution, and Conservation Status, 9th ed. Vol. 8 (Chelonian Research Foundation and Turtle Conservancy, 2021).
Uetz, P., F. P. A. R. & H. J. The Reptile Database. http://www.reptile-database.org/ (2022).
Antonelli, A. et al. Amazonia is the primary source of Neotropical biodiversity. Proc. Natl. Acad. Sci. U.S.A. 115, 6034–6039 (2018).
Mittermeier, R. A., van Dijk, P. P., Rhodin, A. G. J. & Nash, S. D. Turtle hotspots: An analysis of the occurrence of tortoises and freshwater turtles in biodiversity hotspots, high-biodiversity wilderness areas, and turtle priority areas. Chelonian Conserv. Biol. 14, 2–10 (2015).
Cunha, F. A. G., Sampaio, I., Carneiro, J. & Vogt, R. C. A New Species of Amazon Freshwater Toad-Headed Turtle in the Genus Mesoclemmys (Testudines: Pleurodira: Chelidae) from Brazil. Chelonian Conserv. Biol. 20, 151–166 (2021).
Brito, E. S. et al. New records of mesoclemmys raniceps (Testudines, chelidae) for the states of amazonas, pará and Rondônia, north Brazil, including the Tocantins basin. Herpetol. Notes 12, 283–289 (2019).
Cunha, F. A. G. et al. Distribution of Chelus fimbriata and Chelus orinocensis (Testudines: Chelidae). Chelonian Conserv. Biol. 20, 109–115 (2021).
Pritchard, P. Chelus fimbriata (Schneider 1783)—Matamata Turtle. In Conservation Biology of Freshwater Turtles and Tortoises 020.1–020.10 (Chelonian Research Foundation, 2008). https://doi.org/10.3854/crm.5.020.fimbriata.v1.2008.
Vogt, R. C. Tartarugas da Amazônia (2008).
Holmstrom, W. F. Preliminary observations on prey herding in the Matamata turtle, Chelus fimbriatus (Reptilia, Testudines, Chelidae). J. Herpetol. 12, 573 (1978).
Teran, A. F., Vogt, R. C. & de Fatima Soares Gomez, M. Food Habits of an assemblage of five species of turtles in the Rio Guapore, Rondonia, Brazil. J. Herpetol. 29, 536 (1995).
Vargas-Ramírez, M. et al. Genomic analyses reveal two species of the matamata (Testudines: Chelidae: Chelus spp.) and clarify their phylogeography. Mol. Phylogenet. Evol. 148 (2020).
Lasso, C. A. et al. Conservación y tráfico de la tortuga matamata, Chelus fimbriata (Schneider, 1783) en Colombia: un ejemplo del trabajo conjunto entre el Sistema Nacional Ambiental, ONG y academia. Biota Colombiana 19, 147–159 (2018).
Barros, R. M., Sampaio, M. M., Assis, M. F., Ayres, M. & Cunha, O. R. General considerations on the karyotypic evolution of chelonia from the Amazon Region of Brazil. Cytologia 41, 559–565 (1976).
Bull, J. J. & Legler, J. M. Karyotypes of side-necked turtles (Testudines: Pleurodira). Can. J. Zool. 58, 828–841 (1980).
Viana, P. F. et al. An optimized protocol for obtaining mitotic chromosomes from cultured reptilian lymphocytes. Nucleus 59,1–5 (2016).
Mcbee, K., Bickham, J. W., Rhodin, A. G. J. & Mittermeier, R. A. Karyotypic Variation in the Genus Platemys (Testudines: Pleurodira). Copeia 2, 445–449 (1987).
Mazzoleni, S. et al. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Scientific Reports 10, 1–11 (2020).
Viana, P. F. et al. The Amazonian red side-necked turtle Rhinemys rufipes (Spix, 1824) (Testudines, Chelidae) Has a GSD sex-determining mechanism with an ancient XY sex microchromosome system. Cells 9, 1–15 (2020).
Ewert, M. A., Etcheberger, C. R. & Nelson, C. E. Turtle Sex-determining modes and TSD Patterns, and Some TSD Pattern Correlates 21–32 (Smithsonian Books, Washington, 2004).
Ferreira-Júnior Paulo. Aspectos Ecológicos da Determinação Sexual em Tartarugas. 39, 139–154 (2009).
Martinez, P. A., Ezaz, T., Valenzuela, N., Georges, A. & Marshall Graves, J. A. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii: A new piece in the puzzle of sex chromosome evolution in turtles. Chromosom. Res. 16, 815–825 (2008).
Lee, L. S., Montiel, E. E. & Valenzuela, N. Discovery of putative XX/XY male heterogamety in emydura subglobosa turtles exposes a novel trajectory of sex chromosome evolution in emydura. Cytogenet. Genome Res. 158, 160–169 (2019).
Ezaz, T. et al. An XX/XY sex microchromosome system in a freshwater turtle, Chelodina longicollis (Testudines: Chelidae) with genetic sex determination. Chromosom. Res. 14, 139–150 (2006).
van Doorn, G. S. Evolutionary transitions between sex-determining mechanisms: A review of theory. Sex. Dev. 8, 7–19 (2014).
van Doorn, G. S. & Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909–912 (2007).
Beukeboom, L. W. & Perrin, N. The Evolution of Sex Determination (Oxford University Press, Oxford, 2014).
Bachtrog, D. et al. Sex determination: Why so many ways of doing it?. PLoS Biology 12, e1001899 (2014).
Viana, P. F. et al. Landscape of snake’ sex chromosomes evolution spanning 85 MYR reveals ancestry of sequences despite distinct evolutionary trajectories. Sci. Rep. 10, 1–14 (2020).
Gamble, T. et al. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 32, 1296–1309 (2015).
Pennell, M. W., Mank, J. E. & Peichel, C. L. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 27, 3950–3963 (2018).
Valenzuela, N. & Adams, D. C. Chromosome number and sex determination coevolve in turtles. Evolution 65, 1808–1813 (2011).
Sabath, N. et al. Sex determination, longevity, and the birth and death of reptilian species. Ecol. Evol. 6, 5207–5220 (2016).
Literman, R., Burrett, A., Bista, B. & Valenzuela, N. Putative independent evolutionary reversals from genotypic to temperature-dependent sex determination are associated with accelerated evolution of sex-determining genes in turtles. J. Mol. Evol. 86, 11–26 (2018).
Bista, B., Wu, Z., Literman, R. & Valenzuela, N. Thermosensitive sex chromosome dosage compensation in ZZ/ZW softshell turtles, Apalone spinifera. Philos. Trans. R. Soc. B Biol. Sci. 376, 1–14 (2021).
Montiel, E. E., Badenhorst, D., Tamplin, J., Burke, R. L. & Valenzuela, N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 126, 105–113 (2017).
Lee, L., Montiel, E. E., Navarro-Domínguez, B. M. & Valenzuela, N. Chromosomal rearrangements during turtle evolution altered the synteny of genes involved in vertebrate sex determination. Cytogenet. Genome Res. 157, 77–88 (2019).
Bista, B. & Valenzuela, N. Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 11, 1–11 (2020).
Zexian, Z. et al. Diversity of reptile sex chromosome evolution revealed by cytogenetic and linked-read sequencing. bioRxiv (2021).
Cunha, F. A. G., Fernandes, T., Franco, J. & Vogt, R. C. Reproductive biology and hatchling morphology of the amazon toad-headed turtle (Mesoclemmys raniceps) (Testudines: Chelidae), with notes on species morphology and taxonomy of the mesoclemmys group. Chelonian Conserv. Biol. 18, 195 (2019).
Matsubara, K. et al. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 125, 111–123 (2016).
Gorman, G. C. The chromosomes of Reptilia, a cytotaxonomic interpretation. In Cytotaxonomy and Vertebrate Evolution 347–424 (1973).
Reed, K. M. et al. Cytogenetic analysis of the pleurodine turtle Phrynops hogei and its taxonomic implications. Amphibia Reptilia 12, 203–212 (1991).
Cavalcante, M. G. et al. Physical mapping of repetitive DNA suggests 2n reduction in Amazon turtles Podocnemis (Testudines: Podocnemididae). PLoS ONE 13, 1–13 (2018).
Clemente, L. et al. Interstitial telomeric repeats are rare in turtles. Genes 11, 1–18 (2020).
Singchat, W. et al. Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution?. BMC Genomics 19, 1–16 (2018).
Srikulnath, K., Azad, B., Singchat, W. & Ezaz, T. Distribution and amplification of interstitial telomeric sequences (ITSs) in Australian dragon lizards support frequent chromosome fusions in Iguania. PLoS ONE 14, 1–11 (2019).
Abramyan, J., Ezaz, T., Graves, J. A. M. & Koopman, P. Z and W sex chromosomes in the cane toad (Bufo marinus). Chromosom. Res. 17, 1015–1024 (2009).
Born, G. G. & Bertollo, L. A. C. An XX/XY sex chromosome system in a fish species, Hoplias malabaricus, with a polymorphic NOR-bearing × chromosome. Chromosom. Res. 8, 111–118 (2000).
O’Meally, D. et al. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosom. Res. 18, 787–800 (2010).
Ferreira, M., Garcia, C., Matoso, D. A., de Jesus, I. S. & Feldberg, E. A new multiple sex chromosome system X1X1X2X2/X1Y1X2Y2 in siluriformes: Cytogenetic characterization of Bunocephalus coracoideus (Aspredinidae). Genetica 144, 591–599 (2016).
Yano, C. F., Bertollo, L. A. C., Liehr, T., Troy, W. P. & de Cioffi, M. B. W. Chromosome dynamics in Triportheus Species (Characiformes, Triportheidae): An ongoing process narrated by repetitive sequences. J. Hered. 107, 342–348 (2016).
Symonová, R. Integrative rDNAomics-importance of the oldest repetitive fraction of the eukaryote genome. Genes 10, 1–14 (2019).
Kawai, A. et al. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet. Genome Res. 117, 92–102 (2007).
Badenhorst, D., Stanyon, R., Engstrom, T. & Valenzuela, N. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosom. Res. 21, 137–147 (2013).
Viana, P. F. et al. Genomic organization of repetitive DNAs and differentiation of an XX/XY sex chromosome system in the Amazonian Puffer Fish, Colomesus asellus (Tetraodontiformes). Cytogen. Genome Research 153, 1–9 (2018).
Castoe, T. A. et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol. Evol. 3, 641–653 (2011).
Castoe, T. A. et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 12, 1–8 (2011).
Card, D. C. et al. Two low coverage bird genomes and a comparison of reference-guided versus de novo genome assemblies. PLoS ONE 9, e106649 (2014).
Adams, R. H. et al. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 59, 295–310 (2016).
Pearson, C. E. & Sinden, R. R. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile × loci. Biochemistry 35, 5041–5053 (1996).
Chamberlain, N. L., Driver, E. D. & Miesfeld, R. L. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 22, 3181–3186 (1994).
Sandberg, G. Effect of in vitro promoter methylation and CGG repeat expansion on FMR-1 expression. Nucleic Acids Res. 25, 2883–2887 (1997).
Rubinsztein, D. C. et al. Microsatellite evolution—evidence for directionality and variation in rate between species. Nat. Genet. 10, 337–343 (1995).
Eisen, J. A. Mechanistic basis for microsatellite instability. In Microsatellites: Evolution and Applications (eds Goldstein, D. B. & Schlotterer, C.) 34–48 (Oxford University Press, Oxford, 1999).
Payseur, B. A. & Nachman, M. W. Microsatellite variation and recombination rate in the human genome. Genetics 156, 1285–1298 (2000).
Li, Y. C., Korol, A. B., Fahima, T., Beiles, A. & Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Molecular ecology 11, 2453–2465 (2002).
Klintschar, M. et al. Haplotype studies support slippage as the mechanism of germline mutations in short tandem repeats. Electrophoresis 25, 3344–3348 (2004).
Jonika, M., Lo, J. & Blackmon, H. Mode and tempo of microsatellite evolution across 300 million years of insect evolution. Genes 11, 1–15 (2020).
Shedlock, A. M. et al. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc. Natl. Acad. Sci. 104, 2767–2772 (2007).
Ferreira, G. S., Rincón, A. D., Solórzano, A. & Langer, M. C. Review of the fossil matamata turtles: Earliest well-dated record and hypotheses on the origin of their present geographical distribution. Sci. Nat. 103, 1–12 (2016).
Sumner, A. T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75, 304–306 (1972).
Gross, M. C., Schneider, C. H., Valente, G. T., Martins, C. & Feldberg, E. Variability of 18S rDNA locus among Symphysodon fishes: Chromosomal rearrangements. J. Fish Biol. 76, 1117–1127 (2010).
IJdo, J. W., Wells, R. A., Baldini, A. & Reeders, S. T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 19, 4780–4780 (1991).
Viana, P. F. et al. Is the karyotype of neotropical boid snakes really conserved? Cytotaxonomy, chromosomal rearrangements and karyotype organization in the Boidae family. PLoS ONE 11, 1–16 (2016).
Viana, P. F., Ezaz, T., Cioffi, M. D. B., Almeida, B. J. & Feldberg, E. Evolutionary insights of the zw sex chromosomes in snakes: A new chapter added by the amazonian puffing snakes of the genus spilotes. Genes 10, 1–15 (2019).
Zwick, M. S. et al. A rapid procedure for the isolation of C0t−1 DNA from plants. Genome 40, 138–142 (1997).
Ferreira, A. M. V. et al. Cytogenetic Analysis of Panaqolus tankei Cramer & Sousa, 2016 (Siluriformes, Loricariidae), an Ornamental Fish Endemic to Xingu River, Brazil. Cytogenet. Genome Res. 161, 187–194 (2021).
Levan, A., Fredga, K. & Sandberg, A. A. Nomenclature for centromeric position on chromosomes. Hereditas 52, 201–220 (1964).
Acknowledgements
This study is in memory of Richard Carl Vogt, who chose the Amazon as home. Richard was my first supervisor in scientific life, a pioneer in the study of turtles around the world and dedicate his life to studying this charismatic group of reptiles, with great and important scientific contributions to the evolutionary biology of turtles. We are grateful to the Rogério Naiff and Leonardo Matos from the Animal Husbandry of the National Institute of Amazonian Research (Biotério Central—INPA); Milena and Breno Almeida from the Amazonian Center for Herpetology (Centro Amazônico de Herpetologia) for all valuable support provided.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Amazonas (FAPEAM), CNPq Productivity Grants (Process: 301886/2019–9) and Center for Studies on Adaptations of Aquatic Biota of the Amazon (ADAPTA).
Author information
Authors and Affiliations
Contributions
Conceptualization, designed the study and initial structure, P.V., Methodology, P.V.; F.H.T.; S.M.; R.C.V., Validation, P.V.; E.F.; T.E., Formal analysis and investigation, P.V.; E.F.; F.H.T.; S.M.; R.C.V.; T.E. All authors analyzed and interpreted the data. P.V wrote the manuscript with contributions from all coauthors. All coauthors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Viana, P.F., Feldberg, E., Takagui, F.H. et al. Matamatas Chelus spp. (Testudines, Chelidae) have a remarkable evolutionary history of sex chromosomes with a long-term stable XY microchromosome system. Sci Rep 12, 6676 (2022). https://doi.org/10.1038/s41598-022-10782-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10782-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.