Abstract
Tridacna crocea is an ecologically important marine bivalve inhabiting tropical coral reef waters. High quality and available genomic resources will help us understand the population structure and genetic diversity of giant clams. This study reports a high-quality chromosome-scale T. crocea genome sequence of 1.30 Gb, with a scaffold N50 and contig N50 of 56.38 Mb and 1.29 Mb, respectively, which was assembled by combining PacBio long reads and Hi-C sequencing data. Repetitive sequences cover 71.60% of the total length, and a total of 25,440 protein-coding genes were annotated. A total of 1,963 non-coding RNA (ncRNA) were determined in the T. crocea genome, including 62 micro RNA (miRNA), 58 small nuclear RNA (snRNA), 83 ribosomal RNA (rRNA), and 1,760 transfer RNA (tRNA). Phylogenetic analysis revealed that giant clams diverged from oyster about 505.7 Mya during the evolution of bivalves. The genome assembly presented here provides valuable genomic resources to enhance our understanding of the genetic diversity and population structure of giant clams.
Similar content being viewed by others
Background & Summary
Giant clams are tropical marine shellfish mainly distributed in the Indian Ocean, Western Pacific, and South China Sea. There are twelve species of giant clams, divided into two genera, with10 species in Tridacna and 2 in Hippopus1. They play a crucial role in coral reef ecosystems, contributing over 60% of the biomass of coral reef ecosystems2. Giant clams support coral reef biodiversity, offer habitats, breeding and feeding grounds to various marine organisms, and have extremely important ecological value3,4. Giant clams are hermaphrodites, initially functioning as males and later developing female gonads and functioning both as male and female5. To avoid the occurrence of self-fertilization, giant clams first release sperm, and then eggs6. Bivalves often form symbiotic associations with bacteria, algae, and other marine fauna7. There is a symbiotic relationship between giant clams and zooxanthellae. Unlike intracellular symbiosis in stony corals, the zooxanthellae in clams are intercellular and live within the mantle8. The symbionts supply nutrients to the host through photosynthesis. While also obtaining some essential nutrients from the host. Notably, symbionts are not transmitted vertically and must be acquired from the environment during the ontogeny of the second larval stage, veliger9. Additionally, some bivalves from deep sea engage in symbiosis with chemosynthetic bacteria, which are the primary producers of deep-sea cold seeps and vents10.
Among Tridacna species, T. crocea is the smallest, with a maximum shell length of no more than 20 cm, growing at a rate of about 4 cm per year, reaching sexual maturity in 1–2 years11. The shell is shallow, with two equal sides and the same shape and size. Despite its slow growth and small size, T. crocea is known for its vibrant colors and beautiful appearance, making it valuable in food markets, the aquarium trade markets and tropical coral reef ecosystems12. Moreover, its photoautotrophic characteristics contribute to oxygen production, benefiting marine organisms13. However, anthropogenic disturbances, such as global warming, habitat destruction and over-harvesting, have led to declining giant clam populations, resulting in giant clams been listed on the IUCN red list (IUCN, 2007).
Despite the ecological importance of giant clams, their genomic features have remained unclear. In fact, previous molecular studies of giant claims have focused on phylogeographical patterns14,15, as well as the expression and functional analysis of specific genes16,17. Limited transcriptome data are available18,19. Recently, a genomic survey and resources for T. crocea were conducted, which involved determining the genome size, predicting unique content, and providing partial annotations, and assemblies20. The lack of genomic information has been a hindrance to the study of the evolutionary and ecological characteristics of giant clam. Recently, the Pacific Biosciences (PacBio) high-fidelity reads (HiFi) have been successfully applied to various complex species and sex chromosomes, such as cultivated apple (high heterozygous)21, cultivated alfalfa (utotetraploid)22, and human X chromosome23. In the present study, the chromosome-level genome of T. crocea was analyzed for the first time using PacBio HiFi reads, Phase genomics Proximo Hi-C technologies, and Illumina short-read sequencing. In order to predict the relationship between T. crocea and other bivalves, gene prediction, functional annotation and phylogenetic analysis were performed. The genome sequence of the giant clam is an important resource for genetic and breeding studies.
Methods
Experimental samples collection and sequencing
T. crocea were sampled from a tropical marine biological research station in Sanya, Hainan province. The giant clams were immediately anaesthetized, and muscle was extracted for DNA isolation using the modified cetyltrimethylammonium bromide (CTAB) method. The quality and quantity of genomic DNA were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and a Qubit 2.0 fluorometer (Thermo Fisher Scientific). DNA integrity was confirmed using a 0.8% agarose gel.
Three distinct genome libraries were created and sequenced in accordance with the manufacturer’s instructions to produce a chromosome-scale assembly of the giant clam: (i) In accordance with the standard PacBio methodology, PCR-free SMRTbell DNA libraries were created utilizing the BluePippin size selection system. The PacBio Sequel system was used to produce long reads; (ii) Phase Genomic’s Hi-C chromosomal conformation captured reads were prepared with the Proximo Hi-C (Animal) Prep Kit and sequenced; (iii) Purified DNA was sheared using a focused ultrasonicator (Covaris) and then used for 350-bp paired-end library construction with the Next Ultra DNA library prep kit (NEB) for Illumina sequencing, the Illumina NovaSeq. 6000 platform was used to sequence short reads (150 bp in length). RNA was extracted from the giant clam mantle and sequenced on the Illumina NovaSeq platform in order to fully aid gene annotation. To construct a high-quality reference genome for the Tridacna crocea, the whole genome sequencing generated ~167 × Pacbio Sequel long reads (218.24 Gb) (Table 1), ~105 × Hi-C reads (136.70 Gb) and ~45 × Illumina paired-end reads (58.50 Gb) (Table 2).
Genome assembly with Pacbio data and Hi-C data
The Pacbio reads were firstly assembled with Falcon software packages (v2.0.5)24 to build the primary contigs and alternate haplotigs (alternative sequences for regions within the primary contigs where heterozygosity was detectable with the long reads). Tool arrow (v2.2.2) as implemented in SMRTlink6.0 (Pacific Biosciences of California, Inc) was used to polish the contigs. The FALCON-Phase software (v0.2.0-beta) was then used to perform a Hi-C-based contigs phasing, resulting in phased, diploid contigs. The chromosome-scale scaffolds were constructed from the phased contigs using Phase Genomics’ Proximo Hi-C genome scaffolding platform25. Subsequently, Juicebox (v1.8.8)26 was used for a round of polishing to fix minor mistakes in chromosome assignment, ordering, and orientation during chromosomal scaffolding. After a draft set of scaffolds was generated, FALCON-Phase was run again for Hi-C based scaffold phasing. The Illumina sequencing data were further used to improve the assembly by Pilon (v1.22) software27. Finally, the Pacbio reads were initially assembled with Falcon software packages, producing an initial contig assembly, then the assembly was integrated with Phase Genomics Hi-C data to orient and order contigs into chromosome-scale scaffolds. About 78.88% of the 1.30 Gb final Tridacna crocea assembly was assigned to 18 superscaffolds (Fig. 1), with a scaffold N50 and contig N50 of 56.38 Mb and 1.29 Mb, respectively (Table 3). The length distribution of pacbio long reads indicates the peak length is longer than 4 kb (Fig. S1). This result is consistent with the results of other aquatic animals28,29,30,31,32.
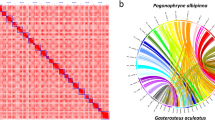
Hi-C contact heatmap and Circos plot illustrate the characterization of Tridacna crocea genome, (A) Genome-wide analysis of chromatin interactions in the Tridacna crocea genome. (B) I: Syntenic regions within the Tridacna crocea assembly base on homology searches carried out by conducting with MCscan (version 0.8) requiring at least 10 genes per block; II: GC content in non-overlapping 1 Mb windows; III: Percent coverage of TEs in non-overlapping 1 Mb windows; IV: Gene density calculated on the basis of the number of genes in non-overlapping 1 Mb windows; V: The length of scaffolds in the size of Mb.
Repeat annotation
There are a large number of repeat sequences in the Tridacna crocea genome, which can be divided into two categories according to the distribution pattern, namely tandem repeat sequences and interspersed repeat sequences. Tandem repetitive sequences were identified using GMATA33 and Tandem Repeats Finder (TRF, version 4.07b)34 with default parameters. The interspersed repeat contents of the Tridacna crocea genome were identified using two methods, de novo repeat identification and known repeat searching against existing databases. RepeatModeler (v1.0.11) and MITE-hunter35 were used to de novo predict repeat sequences in the genome, the homology-based approach involved applying RepeatMasker (version 1.331) (http://www.repeatmasker.org/) and Repbase database36 to identify TE repeats in the assembled genome. The results showed that 71.60% of the assembly consisted of repetitive sequences (Table 4, Fig. 2). The proportion of repeat elements was higher than that of close relatives of mollusks, such as Patinopecten yessoensis (39%)37, Crassostrea gigas (43%)38 and Sinonovacula constricta (40%)29, given that repetitive sequences are the main drivers of genome amplification, T.crocea presents a larger genome size compared to the three closely related species (Table 5). Among these repetitive sequences, transposable elements (TEs) accounted for 55.83% of the T. crocea genome size, with DNA transposons to be the most predominant type (37.68% of the genome size).
Gene prediction and functional annotation
Gene prediction in a repeat-masked genome was performed using reference guided transcriptome assembly, homology search and ab initio prediction. By combining transcriptome alignment, homologous protein prediction and ab initio prediction. In detail, proteins of four mollusks (Crassostrea gigas, Crassostrea virginica, Mizuhopecten yessoensis, Octopus bimaculoides) were downloaded from NCBI DataBase for homolog prediction, GeMoMa39 was used to align the homologous peptides to the assembly and then got the gene structure information. For RNAseq-based gene prediction, filtered mRNA-seq reads were aligned to the reference genome using STAR40. The transcripts were then assembled using StringTie241 and open reading frames (ORFs) were predicted using PASA42. For the de novo prediction, RNA-seq reads were de novo assembled using stringtie and analyzed with PASA to produce a training set. Augustus43 with default parameters were then utilized for ab initio gene prediction with the training set. Finally, EVidenceModeler (EVM)44 was used to produce an integrated gene set of which gene with TE were removed using TransposonPSI package (http://transposonpsi.sourceforge.net/) and the miscoded genes were further filtered. Untranslated regions (UTRs) and alternative splicing regions were determined using PASA based on RNA-seq assemblies. We retained the longest transcripts for each locus, and regions outside of the ORFs were designated UTRs. We predicted 25,440 protein-coding genes with an average gene length of 25,946 bp and an average 8.43 exons per gene. Functional annotation based on public databases (including SwissProt, NR, KEGG, KOG and Gene Ontology) estimated that 23,017 (90.48%) genes could be classified by at least one of the databases (Fig. 3). In addition, we annotated four types of non-coding RNAs in the T. crocea assembly, including micro RNA (miRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), and small nuclear RNA (snRNA). The tRNA genes were predicted by an improved tool for tRNA detection, tRNAscan-SE (version 1.3.1)45 with default paramerters. The rRNA fragments were predicted by aligning to invertebrate template rRNA sequences using BlastN (version 2.2.24) at an E-value of 1e-5. The snRNAs as well as miRNAs were identified using INFERNAL (version 1.1.1)46 to search against the Rfam database (release 12.0). A total of 1,963 non-coding RNA (ncRNA) were determined in the Tridacna crocea genome, including 62 micro RNA (miRNA), 58 small nuclear RNA (snRNA), 83 ribosomal RNA (rRNA), and 1,760 transfer RNA (tRNA) (Table 6).
Intersections of gene families between eleven animals (Tridacna crocea, Aplysia californica, Crassostrea gigas, Crassostrea virginica, Helobdella robusta, Lottia gigantea, Mizuhopecten yessoensis, Octopus bimaculoides, Drosophila melanogaster, Homo sapiens and Nematostella vectensis). The figure was constructed by UpSetR, in which the rows represent the gene families and the columns represent their intersections. Black filled circle represents part of a given intersection; light gray circle represents not a part of the intersection. Bar chart placed on top of the matrix represents the size of the intersections. A second bar chart represents the size of the each set.
Comparative genomic and phylogenetic analysis
We clustered the protein-coding genes into gene families for T. crocea Aplysia californica (GCF_000002075.1), Crassostrea gigas (GCF_902806645.1), Crassostrea virginica (GCF_002022765.2), Helobdella robusta (GCF_000326865.1), Lottia gigantean (GCF_000327385.1), Mizuhopecten yessoensis (GCF_002113885.1), Octopus bimaculoides (GCF_001194135.1), Drosophila melanogaster (GCF_000001215.4), Homo sapiens (GCF_000001405.39) and Nematostella vectensis (GCF_000209225.1) (Table 7). 27,422 gene families were identified, of which 3,109 were shared by all eleven species. Comparing with other ten species, there are 347 specific gene families in the T. crocea assembly (Fig. 3), among these T. crocea specific families, 953 genes are supported by evidence of gene functional annotation. These T. crocea specific genes were significantly (P < 0.05) enriched in zinc ion binding, extracellular ligand-gated ion channel activity, integral component of membrane, ion transport related gene ontology (GO) categories (Table 8).
A phylogenetic tree was constructed using the eleven animal species (Fig. 4). Protein sequences were extracted from each family and concatenated to form one supergene for each species, and the maximum likelihood method47 was used to reconstruct the phylogenetic tree. The divergence time among the eleven animals was estimated using the MCMCtree program (version 4.4) as implemented in the Phylogenetic Analysis of Maximum Likelihood (PAML) package48, with a correlated rates clock and JC69 nucleotide substitution model. The divergence time between T. crocea and M. yessoensis was predicted to be about 505.7 million years ago (MYA). Compared with the common ancestor of T. crocea, M. yessoensis, C. gigas and C. virginica, Tridacna crocea shows 93 and 15 events of gene family expansion and gene family contraction, respectively. The expanded genes in T. crocea are related with “DNA replication” (GO:0006260), “DNA-directed DNA polymerase activity” (GO:0003887), “nucleotide binding” (GO:0000166), “methyltransferase activity” (GO:0008168), and so on. On the other side, the contracted genes in the T. crocea were significantly (P < 0.05) enriched in GO terms for “iron ion binding” (GO:0005506), “heme binding” (GO:0020037), “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen” (GO:0016705), and “oxidation-reduction process” (GO:0055114).
Data Records
The raw Illumina, PacBio, Hi-C sequencing and full length transcriptome data are deposited in the NCBI SRA database under the accession numbers SRR1713764449, SRR1713764550, SRR1713764351, and SRR2565102152, respectively. The genome assembly and annotations are available from the Figshare53,54 and the assembly genome are also deposited at the NCBI with accession number GCA_032873355.155.
Technical Validation
Evaluation of the genome assembly
The Hi-C heatmap exhibits the accuracy of genome assembly, with relatively independent Hi-C signals observed between the 18 pseudo-chromosomes (Fig. 1B). To evaluate the quality of the genome assembly, the completeness of the genome assembly was assessed using the conserved metazoan gene set “metazoan_odb10” from the Benchmarking Universal Single-Copy Orthologs (BUSCO) v4.054. The genome assembly was found to have a high level of completeness (94.2%). 74.2% were complete and single-copy, 20% complete and duplicated, 0.6% fragmented, and 5.2% were missing (Table 9). This demonstrates the remarkable completeness and conservation of gene content in giant clam genome assembly, achieving one of the best BUSCO scores observed among reported mollusks. Therefore, these results suggested that the quality of this genome assembly is high.
Genome annotation and phylogenetic analysis
By comparing with public databases including Gene Ontology, KOG, SwissProt, KEGG and NR, gene function information, motifs and domains of their proteins were assigned (Table 10). InterProScan program56 with default parameters was used to identify the GO terms and putative domains of genes. For other four databases, the EvidenceModeler-integrated protein sequences against the 4 public protein database were compared using BLASTp57 with an E value cutoff of 1e−05. Results from the five database searches were concatenated.
The maximum likelihood method was performed to reconstruct the phylogenetic tree according to47. The divergence time among the eleven animals were predicted by the MCMCtree program (version 4.4) of Phylogenetic Analysis of Maximum Likelihood (PAML) package48, with a correlated rates clock and JC69 nucleotide substitution model. The TimeTree database was used to predict the calibration times of divergence between Octopus bimaculoides and Crassostrea gigas (~554MYA)58.
Code availability
All data processing commands and pipelines are executed according to instructions and guidelines provided by relevant bioinformatics software. No custom scripts or code were used in this study.
References
Neo, M. L., Eckman, W., Vicentuan, K., Teo, S. L. M. & Todd, P. A. The ecological significance of giant clams in coral reef ecosystems. Biol Conserv 181, 111–123, https://doi.org/10.1016/j.biocon.2014.11.004 (2015).
Harzhauser, M., Mandic, O., Piller, W. E., Reuter, M. & Kroh, A. Tracing back the origin of the Indo-Pacific mollusc fauna: Basal Tridacninae from the Oligocene and Miocene of the Sultanate of Oman. Palaeontology 51, 199–213, https://doi.org/10.1111/j.1475-4983.2007.00742.x (2008).
Perry, C. T. et al. Estimating rates of biologically driven coral reef framework production and erosion: a new census-based carbonate budget methodology and applications to the reefs of Bonaire. Coral Reefs 31, 853–868, https://doi.org/10.1007/s00338-012-0901-4 (2012).
Mallela, J. & Perry, C. T. Calcium carbonate budgets for two coral reefs affected by different terrestrial runoff regimes, Rio Bueno, Jamaica. Coral Reefs 26, 129–145, https://doi.org/10.1007/s00338-006-0169-7 (2007).
Mies, M. & Sumida, P. Giant Clam Aquaculture: a Review on Induced Spawning and Larval Rearing. International Journal of Marine Science 2, 62–69 (2012).
Braley, R. D. Serotonin-Induced Spawning in Giant Clams (Bivalvia, Tridacnidae). Aquaculture 47, 321–325, https://doi.org/10.1016/0044-8486(85)90217-0 (1985).
Dubilier, N., Bergin, C. & Lott, C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nature reviews. Microbiology 6, 725–740, https://doi.org/10.1038/nrmicro1992 (2008).
Norton, J., Shepherd, M., Long, H. & Fitt, W. The Zooxanthellal Tubular System in the Giant Clam. Biological Bulletin 183, https://doi.org/10.2307/1542028 (1992).
Mies, M. Evolution, diversity, distribution and the endangered future of the giant clam-Symbiodiniaceae association. Coral Reefs 38, https://doi.org/10.1007/s00338-019-01857-x (2019).
Guo, Y. et al. Hologenome analysis reveals independent evolution to chemosymbiosis by deep-sea bivalves. BMC biology 21, 51, https://doi.org/10.1186/s12915-023-01551-z (2023).
Zhou, Z. et al. Artificial interspecific hybridization of two giant clams, Tridacna squamosa and Tridacna crocea, in the south China sea. Aquaculture 515, 734581, https://doi.org/10.1016/j.aquaculture.2019.734581 (2020).
Li, Y. Q. et al. Study on the Individual Coloring Mechanism of Iridescent Cells in the Mantle of the Boring Giant Clam. Front Mar Sci 9, https://doi.org/10.3389/Fmars.2022.883678 (2022).
Li, J. et al. Assessment of the juvenile vulnerability of symbiont-bearing giant clams to ocean acidification. Sci Total Environ 812, https://doi.org/10.1016/j.scitotenv.2021.152265 (2022).
Cai, S. Y., Mu, W. D., Wang, H., Chen, J. W. & Zhang, H. B. Sequence and phylogenetic analysis of the mitochondrial genome of giant clam, Tridacna crocea (Tridacninae: Tridacna). Mitochondrial DNA B 4, 1032–1033, https://doi.org/10.1080/23802359.2019.1579071 (2019).
Ma, H. T. et al. Molecular phylogeny and divergence time estimates for native giant clams (Cardiidae: Tridacninae) in the Asia-Pacific: Evidence from mitochondrial genomes and nuclear 18S rRNA genes. Front Mar Sci 9, https://doi.org/10.3389/Fmars.2022.964202 (2022).
Zhou, Y. Y. et al. Developmental Expression Pattern of the Piwi1 Gene, Timing of Sex Differentiation and Maturation in Artificially Produced Juvenile Boring Giant Clam, Tridacna crocea. Front Mar Sci 9, https://doi.org/10.3389/Fmars.2022.883661 (2022).
Zhou, Y. et al. Examination of the role of the forkhead box L2 (Foxl2) in gonadal and embryonic development in the boring giant clam Tridacna crocea. Aquaculture 560, 738554, https://doi.org/10.1016/j.aquaculture.2022.738554 (2022).
Zhou, Z., Liu, Z., Wang, L., Luo, J. & Li, H. Oxidative stress, apoptosis activation and symbiosis disruption in giant clam Tridacna crocea under high temperature. Fish & Shellfish Immunology 84, 451–457, https://doi.org/10.1016/j.fsi.2018.10.033 (2019).
Xu, D. et al. Mechanistic molecular responses of the giant clam Tridacna crocea to Vibrio coralliilyticus challenge. Plos One 15, https://doi.org/10.1371/journal.pone.0231399 (2020).
Baeza, J. A., Neo, M. L. & Huang, D. Genomic Survey and Resources for the Boring Giant Clam Tridacna crocea. Genes (Basel) 13, https://doi.org/10.3390/genes13050903 (2022).
Sun, X. P. et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat Genet 52, 1423–1432, https://doi.org/10.1038/s41588-020-00723-9 (2020).
Chen, H. T. et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat Commun 11, https://doi.org/10.1038/S41467-020-16338-X (2020).
Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79-+, https://doi.org/10.1038/s41586-020-2547-7 (2020).
Chin, C. S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13, 1050-+, https://doi.org/10.1038/Nmeth.4035 (2016).
Bickhart, D. M. et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet 49, 643-+, https://doi.org/10.1038/ng.3802 (2017).
Durand, N. C. et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst 3, 99–101, https://doi.org/10.1016/j.cels.2015.07.012 (2016).
Walker, B. J. et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. Plos One 9, https://doi.org/10.1371/journal.pone.0112963 (2014).
Gomes-dos-Santos, A. et al. The Crown Pearl: a draft genome assembly of the European freshwater pearl mussel Margaritifera margaritifera (Linnaeus, 1758). DNA Research 28, https://doi.org/10.1093/dnares/dsab002 (2021).
Ran, Z. et al. Chromosome-level genome assembly of the razor clam Sinonovacula constricta (Lamarck, 1818). Mol Ecol Resour 19, 1647–1658, https://doi.org/10.1111/1755-0998.13086 (2019).
Gallardo-Escarate, C. et al. Chromosome-Level Genome Assembly of the Blue Mussel Mytilus chilensis Reveals Molecular Signatures Facing the Marine Environment. Genes-Basel 14, https://doi.org/10.3390/Genes14040876 (2023).
Bai, C.-M. et al. Chromosomal-level assembly of the blood clam, Scapharca (Anadara) broughtonii, using long sequence reads and Hi-C. Gigascience 8, https://doi.org/10.1093/gigascience/giz067 (2019).
Kim, J. et al. Chromosome-Level Genome Assembly of the Butter Clam Saxidomus purpuratus. Genome Biology and Evolution 14, https://doi.org/10.1093/gbe/evac106 (2022).
Wang, X. W. & Wang, L. GMATA: An Integrated Software Package for Genome-Scale SSR Mining, Marker Development and VIewing. Front Plant Sci 7, https://doi.org/10.3389/Fpls.2016.01350 (2016).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580, https://doi.org/10.1093/Nar/27.2.573 (1999).
Han, Y. J. & Wessler, S. R. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res 38, https://doi.org/10.1093/nar/gkq862 (2010).
Bao, W. D., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA-Uk 6, https://doi.org/10.1186/s13100-015-0041-9 (2015).
Wang, S. et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol 1, 120, https://doi.org/10.1038/s41559-017-0120 (2017).
Penaloza, C. et al. A chromosome-level genome assembly for the Pacific oyster Crassostrea gigas. Gigascience 10, https://doi.org/10.1093/gigascience/giab020 (2021).
Keilwagen, J. et al. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res 44, https://doi.org/10.1093/nar/gkw092 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol 15, https://doi.org/10.1186/S13059-014-0550-8 (2014).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol 20, https://doi.org/10.1186/S13059-019-1910-1 (2019).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31, 5654–5666, https://doi.org/10.1093/nar/gkg770 (2003).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34, W435–W439, https://doi.org/10.1093/nar/gkl200 (2006).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 9, https://doi.org/10.1186/Gb-2008-9-1-R7 (2008).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964, https://doi.org/10.1093/Nar/25.5.955 (1997).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935, https://doi.org/10.1093/bioinformatics/btt509 (2013).
Guindon, S. et al. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol 59, 307–321, https://doi.org/10.1093/sysbio/syq010 (2010).
Yang, Z. H. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13, 555–556 (1997).
Li, J. NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR17137644 (2023).
Li, J. NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR17137645 (2023).
Li, J. NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR17137643 (2023).
Li, J. NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRR25651021 (2023).
Li, J. Chromosome-level genome assembly and annotation of rare and endangered tropical bivalve, Tridacna crocea. figshare. Dataset. https://doi.org/10.6084/m9.figshare.24264643 (2023).
Li, J. Chromosome-level genome assembly and annotation of rare and endangered tropical bivalve, Tridacna crocea. figshare. Dataset. https://doi.org/10.6084/m9.figshare.24264646 (2023).
Li, J. NCBI Genbank https://identifiers.org/insdc.gca:GCA_032873355.1 (2023).
Quevillon, E. et al. InterProScan: protein domains identifier. Nucleic Acids Res 33, W116–W120, https://doi.org/10.1093/nar/gki442 (2005).
McGinnis, S. & Madden, T. L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32, W20–W25, https://doi.org/10.1093/nar/gkh435 (2004).
Hedges, S. B. & Dudley, J. Kumar & Sudhir. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (2006).
Acknowledgements
This research was supported by National Key Research and Development Program of China (2022YFC3102002); Guangzhou Science and Technology Project (2023B03J00165; 202206010133); Guangdong Provincial Key Research and Development Program (2021B0202020003); the National Science Foundation of China (32002387); the Project of Sanya Yazhou Bay Science and Technology City; Science and Technology Project of Guangdong Provincial Department of Natural Resources (GDNRC[2022]40); Guangdong Basic and Applied Basic Research Foundation (2023A1515010944; 2022A1515010203); the Open Foundation of the State Key Laboratory of Loess and Quaternary Geology (SKLLQG2213); National Marine Genetic Resource Center; the earmarked fund for CARS-49; and the Science and Technology Planning Project of Guangdong Province, China (2023B1212060047).
Author information
Authors and Affiliations
Contributions
Jun Li, Yuehuan Zhang, and Ziniu Yu conceived and designed this study. Yanping Qin and Haitao Ma collected the samples. Yongchao Niu and Zhen Zhao assembled and annotated the genome. Yongchao Niu and Zhen Zhao performed gene family and genome evolutionary analyses. Jun Li, Yanping Qin, Jianmin Lian, Zohaib Noor, Shuming Guo, Gongpengyang Shi and Jiang Li performed bioinformatic analyses. Jun Li wrote the manuscript. Yanping Qin, and Haitao Ma revised it. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Ma, H., Qin, Y. et al. Chromosome-level genome assembly and annotation of rare and endangered tropical bivalve, Tridacna crocea. Sci Data 11, 186 (2024). https://doi.org/10.1038/s41597-024-03014-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03014-8