Abstract
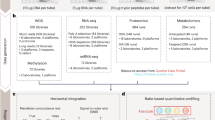
The growing number of multi-omics studies demands clear conceptual workflows coupled with easy-to-use software tools to facilitate data analysis and interpretation. This protocol covers three key components involved in multi-omics analysis, including single-omics data analysis, knowledge-driven integration using biological networks and data-driven integration through joint dimensionality reduction. Using the dataset from a recent multi-omics study of human pancreatic islet tissue and plasma samples, the first section introduces how to perform transcriptomics/proteomics data analysis using ExpressAnalyst and lipidomics data analysis using MetaboAnalyst. On the basis of significant features detected in these workflows, the second section demonstrates how to perform knowledge-driven integration using OmicsNet. The last section illustrates how to perform data-driven integration from the normalized omics data and metadata using OmicsAnalyst. The complete protocol can be executed in ~2 h. Compared with other available options for multi-omics integration, the Analyst software suite described in this protocol enables researchers to perform a wide range of omics data analysis tasks via a user-friendly web interface.
Key points
-
This protocol for web-based multi-omics integration covers single-omics data analysis using ExpressAnalyst and MetaboAnalyst, followed by knowledge-driven integration using OmicsNet and data-driven integration using OmicsAnalyst.
-
This series of web-based tools allows researchers to perform a wide range of omics data analysis tasks via a user-friendly web interface, helping to democratize omics data analysis and empower researchers without strong statistics and programming backgrounds.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All datasets used in this protocol are from a previously published study that collected multi-omics data from islet tissue (transcriptomics and proteomics) and plasma (lipidomics) from human donors undergoing pancreatic surgery30. The example datasets used in this protocol are integrated as examples throughout the four web tools. The unprocessed matrices are included as examples in ExpressAnalyst (www.expressanalyst.ca) and MetaboAnalyst (www.metaboanalyst.ca). Lists of significant features are included as examples in OmicsNet (www.omicsnet.ca). Normalized matrices are included as examples in OmicsAnalyst (www.omicsanalyst.ca). See the ‘Materials’ section for further information about the example datasets.
Code availability
ExpressAnalyst, MetaboAnalyst, OmicsNet and OmicsAnalyst are all freely available as web-based applications. The underlying R packages for each tool are freely available as GitHub repositories: ExpressAnalystR (https://github.com/xia-lab/ExpressAnalystR), MetaboAnalystR (https://github.com/xia-lab/MetaboAnalystR), OmicsNetR (https://github.com/xia-lab/OmicsNetR) and OmicsAnalystR (https://github.com/xia-lab/OmicsAnalystR) under the GNU General Public License version 2 or later.
References
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol. 18, 83 (2017).
Han, J.-D. J. Understanding biological functions through molecular networks. Cell Res. 18, 224–237 (2008).
Subramanian, I., Verma, S., Kumar, S., Jere, A. & Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 14, 1177932219899051 (2020).
Eicher, T. et al. Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites https://doi.org/10.3390/metabo10050202 (2020).
Huang, S., Chaudhary, K. & Garmire, L. X. More is better: recent progress in multi-omics data integration methods. Front. Genetics https://doi.org/10.3389/fgene.2017.00084 (2017).
Quinn, T. P. et al. A field guide for the compositional analysis of any-omics data. GigaScience https://doi.org/10.1093/gigascience/giz107 (2019).
Verheijen, M. et al. Towards the development of an omics data analysis framework. Reg. Toxicol. Pharmacol. 112, 104621 (2020).
Tarazona, S., Arzalluz-Luque, A. & Conesa, A. Undisclosed, unmet and neglected challenges in multi-omics studies. Nat. Comput. Sci. 1, 395–402 (2021).
Gomez-Cabrero, D. et al. Data integration in the era of omics: current and future challenges. BMC Syst. Biol. 8, I1 (2014).
Palsson, B. & Zengler, K. The challenges of integrating multi-omic data sets. Nat. Chem. Biol. 6, 787–789 (2010).
Jendoubi, T. Approaches to integrating metabolomics and multi-omics data: a primer. Metabolites 11, 184 (2021).
Kim, D. et al. Knowledge boosting: a graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J. Am. Med. Inform. Assoc. 22, 109–120 (2014).
Zhou, G., Li, S. & Xia, J. in Computational Methods and Data Analysis for Metabolomics (ed Li. S) 469–487 (Springer, 2020).
Blatti, C. III et al. Knowledge-guided analysis of “omics” data using the KnowEnG cloud platform. PLoS Biol. 18, e3000583 (2020).
Liu, T. et al. PaintOmics 4: new tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. 50, W551–W559 (2022).
Zhou, G., Pang, Z., Lu, Y., Ewald, J. & Xia, J. OmicsNet 2.0: a web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 50, W527–W533 (2022).
Cantini, L. et al. Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nat. Commun. 12, 124 (2021).
Picard, M., Scott-Boyer, M.-P., Bodein, A., Périn, O. & Droit, A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 19, 3735–3746 (2021).
Argelaguet, R. et al. Multi-Omics Factor Analysis—a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124 (2018).
Rohart, F., Gautier, B., Singh, A. & Lê Cao, K.-A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752 (2017).
McCabe, S. D., Lin, D.-Y. & Love, M. I. Consistency and overfitting of multi-omics methods on experimental data. Brief. Bioinform. 21, 1277–1284 (2019).
Pang, Z. et al. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
Xia, J. & Wishart, D. S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 6, 743–760 (2011).
Chong, J., Liu, P., Zhou, G. & Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821 (2020).
Xia, J., Gill, E. E. & Hancock, R. E. W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 10, 823–844 (2015).
Chang, L. & Xia, J. in Transcription Factor Regulatory Networks (eds Song, Q. & Tao, Z.) 185–204 (Springer, 2023).
Chang, L., Zhou, G., Soufan, O. & Xia, J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48, W244–W251 (2020).
Zhou, G., Ewald, J. & Xia, J. OmicsAnalyst: a comprehensive web-based platform for visual analytics of multi-omics data. Nucleic Acids Res. 49, W476–W482 (2021).
Zhou, G. & Xia, J. OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic Acids Res. 46, W514–W522 (2018).
Wigger, L. et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat. Metab. 3, 1017–1031 (2021).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016).
Pang, Z., Chong, J., Li, S. & Xia, J. MetaboAnalystR 3.0: toward an optimized workflow for global metabolomics. Metabolites 10, 186 (2020).
Röst, H. L. et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741–748 (2016).
Schmid, R. et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 41, 447–449 (2023).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Li, S., Siddiqa, A., Thapa, M., Chi, Y. & Zheng, S. Trackable and scalable LC–MS metabolomics data processing using asari. Nat. Commun. 14, 4113 (2023).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Li, S. et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 9, e1003123 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Xia, J. & Wishart, D. S. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 38, W71–W77 (2010).
Xia, J. & Wishart, D. S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26, 2342–2344 (2010).
Schadt, E. E. Molecular networks as sensors and drivers of common human diseases. Nature 461, 218–223 (2009).
Dianati, N. Unwinding the hairball graph: Pruning algorithms for weighted complex networks. Phys. Rev. E 93, 012304 (2016).
Lovino, M. et al. A survey on data integration for multi-omics sample clustering. Neurocomputing 488, 494–508 (2022).
Meng, C. et al. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinform. 17, 628–641 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biol. 15, 1–21 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Ding, J. et al. Mergeomics 2.0: a web server for multi-omics data integration to elucidate disease networks and predict therapeutics. Nucleic Acids Res. 49, W375–W387 (2021).
Hernández-de-Diego, R. et al. PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res. 46, W503–W509 (2018).
Kuo, T.-C., Tian, T.-F. & Tseng, Y. J. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst. Biol. 7, 64 (2013).
Zoppi, J., Guillaume, J.-F., Neunlist, M. & Chaffron, S. MiBiOmics: an interactive web application for multi-omics data exploration and integration. BMC Bioinform. 22, 6 (2021).
Mirza, B. et al. Machine learning and integrative analysis of biomedical big data. Genes 10, 87 (2019).
Asada, K. et al. Integrated analysis of whole genome and epigenome data using machine learning technology: toward the establishment of precision oncology. Front. Oncol. https://doi.org/10.3389/fonc.2021.666937 (2021).
Cazaly, E. et al. Making sense of the epigenome using data integration approaches. Front. Pharmacol. https://doi.org/10.3389/fphar.2019.00126 (2019).
Simovski, B. et al. GSuite HyperBrowser: integrative analysis of dataset collections across the genome and epigenome. GigaScience https://doi.org/10.1093/gigascience/gix032 (2017).
Kang, M., Ko, E. & Mersha, T. B. A roadmap for multi-omics data integration using deep learning. Brief. Bioinform. https://doi.org/10.1093/bib/bbab454 (2022).
Lee, J., Hyeon, D. Y. & Hwang, D. Single-cell multiomics: technologies and data analysis methods. Exp. Mol. Med. 52, 1428–1442 (2020).
Macaulay, I. C., Ponting, C. P. & Voet, T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 33, 155–168 (2017).
Tarazona, S. et al. Harmonization of quality metrics and power calculation in multi-omic studies. Nat. Commun. 11, 3092 (2020).
de Souza, N. The ENCODE project. Nat. Methods 9, 1046–1046 (2012).
Lonsdale, J. et al. The Genotype–Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Papatheodorou, I. et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 46, D246–D251 (2017).
Gertsman, I. & Barshop, B. A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metab. Dis. 41, 355–366 (2018).
Liu, P. et al. Ultrafast functional profiling of RNA-seq data for nonmodel organisms. Genome Res. 31, 713–720 (2021).
Liu, P. et al. ExpressAnalyst: a unified platform for RNA-sequencing analysis in non-model species. Nat. Commun. 14, 2995 (2023).
Bourgon, R., Gentleman, R. & Huber, W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl Acad. Sci. USA 107, 9546–9551 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Välikangas, T., Suomi, T. & Elo, L. L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 19, 1–11 (2016).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–d368 (2017).
Varlet, A. A. et al. Fine-tuning of actin dynamics by the HSPB8–BAG3 chaperone complex facilitates cytokinesis and contributes to its impact on cell division. Cell Stress Chaperones 22, 553–567 (2017).
Nevins, A. K. & Thurmond, D. C. Glucose regulates the cortical actin network through modulation of Cdc42 cycling to stimulate insulin secretion. Am. J. Physiol. Cell Physiol. 285, C698–C710 (2003).
Belosludtsev, K. N. et al. Alisporivir treatment alleviates mitochondrial dysfunction in the skeletal muscles of C57BL/6NCrl mice with high-fat diet/streptozotocin-induced diabetes mellitus. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22179524 (2021).
Neuhausen, S. L. et al. Genetic variation in insulin-like growth factor signaling genes and breast cancer risk among BRCA1 and BRCA2 carriers. Breast Cancer Res. 11, R76 (2009).
Rosvall, M. & Bergstrom, C. T. Maps of random walks on complex networks reveal community structure. Proc. Natl Acad. Sci. USA 105, 1118–1123 (2008).
Brunk, E. et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 36, 272–281 (2018).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–d386 (2018).
Heslegrave, A. J. et al. Leucine-sensitive hyperinsulinaemic hypoglycaemia in patients with loss of function mutations in 3-hydroxyacyl-CoA dehydrogenase. Orphanet. J. Rare Dis. 7, 25 (2012).
Zhang, W. & Sang, Y. M. Genetic pathogenesis, diagnosis, and treatment of short-chain 3-hydroxyacyl-coenzyme A dehydrogenase hyperinsulinism. Orphanet. J. Rare Dis. 16, 467 (2021).
Gerst, F. et al. The expression of aldolase B in islets is negatively associated with insulin secretion in humans. J. Clin. Endocrinol. Metab. 103, 4373–4383 (2018).
Son, J. et al. Genetic and pharmacologic inhibition of ALDH1A3 as a treatment of β-cell failure. Nat. Commun. 14, 558 (2023).
Warde-Farley, D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220 (2010).
Sun, Y. V. & Hu, Y. J. Integrative analysis of multi-omics data for discovery and functional studies of complex human diseases. Adv. Genet. 93, 147–190 (2016).
Pinu, F. R. et al. Systems biology and multi-omics integration: viewpoints from the metabolomics research community. Metabolites 9, 76 (2019).
Acknowledgements
We thank the Canadian Institutes of Health Research, the Juvenile Diabetes Research Foundation of Canada, Diabetes Canada, the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs Program for funding support.
Author information
Authors and Affiliations
Contributions
J.D.E. and J.X. prepared the manuscript. J.D.E., G.Z., Y.L. and J.X. contributed to the development of the tools (MetaboAnalyst, ExpressAnalyst, OmicsNet and OmicsAnalyst). J.K., C.E., J.D.J. and P.E.M. validated the tools and protocol steps, resulting in improvements to both based on their feedback. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
J.D.E., G.Z. and J.X. own shares of OmicSquare Analytics Inc.
Peer review
Peer review information
Nature Protocols thanks Peter A. C. ‘t Hoen and Sonia Tarazona for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Zhou, G. et al. Nucleic Acids Res. 50, W527–W533 (2022): https://doi.org/10.1093/nar/gkac376
Zhou, G. et al. Nucleic Acids Res. 49, W476–W482 (2021): https://doi.org/10.1093/nar/gkab394
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ewald, J.D., Zhou, G., Lu, Y. et al. Web-based multi-omics integration using the Analyst software suite. Nat Protoc (2024). https://doi.org/10.1038/s41596-023-00950-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-023-00950-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.