Abstract
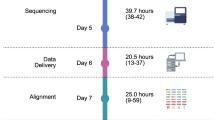
Two decades after the genomics revolution, oncology is rapidly transforming into a genome-driven discipline, yet routine cancer diagnostics is still mainly microscopy based, except for tumor type-specific predictive molecular tests. Pathology laboratories struggle to quickly validate and adopt biomarkers identified by genomics studies of new targeted therapies. Consequently, clinical implementation of newly approved biomarkers suffers substantial delays, leading to unequal patient access to these therapies. Whole-genome sequencing (WGS) can successfully address these challenges by providing a stable molecular diagnostic platform that allows detection of a multitude of genomic alterations in a single cost-efficient assay and facilitating rapid implementation, as well as by the development of new genomic biomarkers. Recently, the Whole-genome sequencing Implementation in standard Diagnostics for Every cancer patient (WIDE) study demonstrated that WGS is a feasible and clinically valid technique in routine clinical practice with a turnaround time of 11 workdays. As a result, WGS was successfully implemented at the Netherlands Cancer Institute as part of routine diagnostics in January 2021. The success of implementing WGS has relied on adhering to a comprehensive protocol including recording patient information, sample collection, shipment and storage logistics, sequencing data interpretation and reporting, integration into clinical decision-making and data usage. This protocol describes the use of fresh-frozen samples that are necessary for WGS but can be challenging to implement in pathology laboratories accustomed to using formalin-fixed paraffin-embedded samples. In addition, the protocol outlines key considerations to guide uptake of WGS in routine clinical care in hospitals worldwide.
Key points
-
Whole-genome sequencing analysis detects DNA changes in the whole cancer genome, allowing identification of new, clinically relevant biomarkers, and opening the doors to a new era of tumor diagnostic and personalized cancer medicine.
-
The protocol provides a detailed workflow covering patient selection, sample handling, and interpretation and reporting of the results, facilitating implementation of whole-genome sequencing analysis in routine oncology clinical practice.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
All code used is open source and available from third parties or developed by Hartwig Medical Foundation (https://github.com/hartwigmedical/). Multiple versions of the pipeline were used during this research (v5.23 and up). Versions of the pipeline are validated and verified according to the Dutch ISO-17025 certification and can be found at https://github.com/hartwigmedical/pipeline5/tags with no restrictions. A schematic overview of the pipeline can be found in Supplementary Fig. 1.
References
Steeghs, E. M. P. et al. Mutation-tailored treatment selection in non-small cell lung cancer patients in daily clinical practice. Lung Cancer 167, 87–97 (2022).
Pasmans, C. T. B. et al. Micro-costing diagnostics in oncology: from single-gene testing to whole- genome sequencing. Expert Rev. Pharmacoecon. Outcomes Res. 21, 413–414 (2021).
Lynch, J. A. et al. Underutilization and disparities in access to EGFR testing among Medicare patients with lung cancer from 2010–2013. BMC Cancer 18, 306 (2018).
Samsom, K. G. et al. Feasibility of whole-genome sequencing-based tumor diagnostics in routine pathology practice. J. Pathol. 258, 179–188 (2022).
Brittain, H. K., Scott, R. & Thomas, E. The rise of the genome and personalised medicine. Clin. Med. 17, 545––551 (2017).
Nakagawa, H. & Fujita, M. Whole-genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 109, 513–522 (2018).
van der Velden, D. L. et al. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature 574, 127–131 (2019).
Schipper, L. J. et al. Clinical impact of prospective whole genome sequencing in sarcoma patients. Cancers (Basel) 14, https://doi.org/10.3390/cancers14020436 (2022).
Schipper, L. J. et al. Complete genomic characterization in patients with cancer of unknown primary origin in routine diagnostics. ESMO Open https://doi.org/10.1016/j.esmoop.2022.100611 (2022).
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Zhang, J. et al. The international cancer genome consortium data portal. Nat. Biotechnol. 37, 367–369 (2019).
Marshall, C. R. et al. The Medical Genome Initiative: moving whole-genome sequencing for rare disease diagnosis to the clinic. Genome Med. 12, 48 (2020).
van de Ven, M. et al. Whole-genome sequencing in oncology: using scenario drafting to explore future developments. BMC Cancer 21, 488 (2021).
Kawanishi, M., Matsuda, T. & Yagi, T. Genotoxicity of formaldehyde: molecular basis of DNA damage and mutation. Front. Environ. Sci. https://doi.org/10.3389/fenvs.2014.00036 (2014).
Bass, B. P., Engel, K. B., Greytak, S. R. & Moore, H. M. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen. Arch. Pathol. Lab. Med. 138, 1520–1530 (2014).
Robbe, P. et al. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genet. Med. 20, 1196–1205 (2018).
Turnbull, C. Introducing whole-genome sequencing into routine cancer care: the Genomics England 100,000 Genomes Project. Ann. Oncol. 29, 784–787 (2018).
Meggendorfer, M. et al. Analytical demands to use whole-genome sequencing in precision oncology. Semin. Cancer Biol. 84, 16–22 (2022).
Roepman, P. et al. Clinical validation of whole genome sequencing for cancer diagnostics. J. Mol. Diagn. 23, 816–833 (2021).
Mandelker, D. et al. Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann. Oncol. 30, 1221–1231 (2019).
VKGN. 20211006 Adviezen projectgroep tumor—en erfelijkheidsdiagnostiek definitief. https://www.artsengenetica.nl/sites/default/files/inline-files/tabel-3-versie-mei-2023.pdf (2021).
Nguyen, L., Van Hoeck, A. & Cuppen, E. Machine learning-based tissue of origin classification for cancer of unknown primary diagnostics using genome-wide mutation features. Nat. Commun. 13, 4013 (2022).
Martínez-Jiménez, F. et al. Genetic immune escape landscape in primary and metastatic cancer. Nat. Genet. 55, 820–831 (2023).
Zhao, C. et al. TruSight Oncology 500: enabling comprehensive genomic profiling and biomarker reporting with targeted sequencing. Preprint at bioRxiv https://doi.org/10.1101/2020.10.21.349100 (2020).
Cheng, D. T. et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Vestergaard, L. K., Oliveira, D. N. P., Poulsen, T. S., Hogdall, C. K. & Hogdall, E. V. Oncomine comprehensive assay v3 vs. Oncomine comprehensive assay plus. Cancers https://doi.org/10.3390/cancers13205230 (2021).
Hesselink, A. T. et al. Clinical validation of Anyplex II HPV HR detection according to the guidelines for HPV test requirements for cervical cancer screening. J. Clin. Virol. 76, 36–39 (2016).
Willems, A. J. et al. Loss of heterozygosity at the BRCA2 locus detected by multiplex ligation-dependent probe amplification is common in prostate cancers from men with a germline BRCA2 mutation. Clin. Cancer Res. 14, 2953–2961 (2008).
van Wijk, L. M., Nilas, A. B., Vrieling, H. & Vreeswijk, M. P. G. RAD51 as a functional biomarker for homologous recombination deficiency in cancer: a promising addition to the HRD toolbox? Expert Rev. Mol. Diagn. 22, 185–199 (2022).
Schouten, P. C. et al. Ovarian cancer-specific BRCA-like copy-number aberration classifiers detect mutations associated with homologous recombination deficiency in the AGO-TR1 trial. Clin. Cancer Res. 27, 6559–6569 (2021).
Szilagyi, M. et al. Circulating cell-free nucleic acids: main characteristics and clinical application. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21186827 (2020).
Wrzeszczynski, K. O. et al. Analytical validation of clinical whole-genome and transcriptome sequencing of patient-derived tumors for reporting targetable variants in cancer. J. Mol. Diagn. 20, 822–835 (2018).
Nelson, A. C. & Yohe, S. L. Cancer whole-genome sequencing: the quest for comprehensive genomic profiling in routine oncology care. J. Mol. Diagn. 23, 784–787 (2021).
Cuppen, E. et al. Implementation of whole-genome and transcriptome sequencing into clinical cancer care. JCO Precis. Oncol. 6, e2200245 (2022).
Hu, B., Sajid, M., Lv, R., Liu, L. & Sun, C. A review of spatial profiling technologies for characterizing the tumor microenvironment in immuno-oncology. Front. Immunol. 13, 996721 (2022).
Laroche, F. J. F. et al. S1P1 Threonine 236 phosphorylation mediates the invasiveness of triple-negative breast cancer and sensitivity to FTY720. Cells https://doi.org/10.3390/cells12070980 (2023).
Hernandez, S. et al. Differential spatial gene and protein expression associated with recurrence following chemoradiation for localized anal squamous cell cancer. Cancers https://doi.org/10.3390/cancers15061701 (2023).
Friedrich, C. et al. Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nat. Commun. 12, 3576 (2021).
Norton, S. E., Lechner, J. M., Williams, T. & Fernando, M. R. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin. Biochem. 46, 1561–1565 (2013).
Orchard, G. E. et al. New embedding and staining systems PrestoCHILL and Presto stainer for application in the advancement of Mohs micrographic surgery. Br. J. Biomed. Sci. 74, 203–208 (2017).
Samsom, K. G. et al. Study protocol: whole-genome sequencing Implementation in standard Diagnostics for Every cancer patient (WIDE). BMC Med. Genomics 13, 169 (2020).
Veneroni, S. et al. Applicability of under vacuum fresh tissue sealing and cooling to omics analysis of tumor tissues. Biopreserv. Biobank 14, 480–490 (2016).
Piaton, E. et al. Guidelines for May-Grunwald-Giemsa staining in haematology and non-gynaecological cytopathology: recommendations of the French Society of Clinical Cytology (SFCC) and of the French Association for Quality Assurance in Anatomic and Cytologic Pathology (AFAQAP). Cytopathology 27, 359–368 (2016).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019).
Kohsaka, S., Petronczki, M., Solca, F. & Maemondo, M. Tumor clonality and resistance mechanisms in EGFR mutation-positive non-small-cell lung cancer: implications for therapeutic sequencing. Future Oncol. 15, 637–652 (2019).
Casparie, M. et al. Pathology databanking and biobanking in the Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 29, 19–24 (2007).
Acknowledgements
We acknowledge ZonMw (the Netherlands Organization for Health Research and Development) Hartwig Medical Foundation and Illumina for funding the original study on which this protocol is based4.
Author information
Authors and Affiliations
Contributions
Designed the protocol: K.G.S., L.J.W.B., L.J.S., D.S., P.R., M.C.B., A.J.d.L., E.d.B., H.V.S., L.E.v.d.K., E.C., E.E.V., J.G.v.d.B., T.E.B., F.L., B.M.H.v.L., K.v.D., K.v.d.B., U.U., L.S., E.G.K., G.A.M. and K.M. Performed experiments: D.S., B.M.H.v.L., E.d.B. and I.R. Interpretation of sequencing results: K.G.S., L.J.W.B., L.J.S., P.R., M.C.B., E.H.R., R.K., F.B.L.H., E.C. and K.M. Participated in multidisciplinary clinical decision-making: K.G.S., L.J.S., L.J.W.B., P.R., M.C.B., A.J.d.L., T.E.B., E.H.R., R.K., J.J.M.v.d.H., L.E.v.d.K. and K.M. Funding: K.M., L.J.W.B., H.V.S., E.C., E.E.V., G.A.M. and J.G.v.d.B. Bioinformatics support: L.S. and D.v.B. Radiology support: F.L. and E.G.K. Logistics design: K.G.S., L.J.S., L.J.W.B., D.S., K.M., K.v.D. and I.R. Wrote the manuscript: K.G.S., L.J.W.B., K.M. and G.A.M. Created figures: K.G.S., E.C., D.S. and L.J.W.B. All authors have read, critically revised and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.J.d.L. reports grants from Bristol Myers Squibb (BMS), Merck Sharp & Dohme (MSD), AstraZeneca and Boehringer, nonfinancial support from Merck Serono and Roche. H.v.S. and E.C. report consultancy fees and support for attending meetings and travelling from Illumina. E.E.V. is a member of the supervisory board of Hartwig. G.A.M. is co-founder and board member (CSO) of CRCbioscreen, he has a research collaboration with CZ Health Insurances (cash matching to ZonMW grant) and has research collaborations with Exact Sciences, Sysmex, Sentinel Ch. SpA, Personal Genome Diagnostics and DELFi; these companies provide materials, equipment and/or sample/genomic analyses. G.A.M. is an advisory board member of ‘Missie Tumor Onbekend’. K.M. reports research grants from AstraZeneca and speakers’ fees from MSD, Roche, AstraZeneca and Benecke. K.M. received consultancy fees from Pfizer, BMS, Roche, MSD, Abbvie, AstraZeneca, Diaceutics, Lilly, Bayer, Boehringer and Ingelheim and nonfinancial support from Roche, Takeda, Pfizer, PGDx and DELFi. The remaining authors report no conflicts of interest.
Peer review
Peer review information
Nature Protocols thanks Hidewaki Nakagawa and Stephen Yip for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Samsom, K. G. et al. J. Pathol. 258, 179–188 (2022): https://doi.org/10.1002/path.5988
van der Velden, D. L. et al. Nature 574, 127–131 (2019): https://doi.org/10.1038/s41586-019-1600-x
van de Haar, J. et al. Nat. Med. 27, 1553–1563 (2021): https://doi.org/10.1038/s41591-021-01448-w
Supplementary information
Supplementary Information
Supplementary Procedure (Parts 9 and 10), Figs. 1 and 2 and Results.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Samsom, K.G., Bosch, L.J.W., Schipper, L.J. et al. Optimized whole-genome sequencing workflow for tumor diagnostics in routine pathology practice. Nat Protoc 19, 700–726 (2024). https://doi.org/10.1038/s41596-023-00933-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00933-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.