Abstract
The isolation of proteins in high yield and purity is a major bottleneck for the analysis of their three-dimensional structure, function and interactome. Here, we present a streamlined workflow for the rapid production of proteins or protein complexes using lentiviral transduction of human suspension cells, combined with highly specific nanobody-mediated purification and proteolytic elution. Application of the method requires prior generation of a plasmid coding for a protein of interest (POI) fused to an N- or C-terminal GFP or ALFA peptide tag using a lentiviral plasmid toolkit we have designed. The plasmid is then used to generate human suspension cell lines stably expressing the tagged fusion protein by lentiviral transduction. By leveraging the picomolar affinity of the GFP and ALFA nanobodies for their respective tags, the POI can be specifically captured from the resulting cell lysate even when expressed at low levels and under a variety of conditions, including detergents and mild denaturants. Finally, rapid and specific elution of the POI (in its tagged or untagged form) under native conditions is achieved within minutes at 4 °C, using the engineered SUMO protease SENPEuB. We demonstrate the wide applicability of the method by purifying multiple challenging soluble and membrane protein complexes to high purity from human cells. Our strategy is also directly compatible with many widely used GFP-expression plasmids, cell lines and transgenic model organisms. Finally, our method is faster than alternative approaches, requiring only 8 d from plasmid to purified protein, and results in substantially improved yields and purity.
Key points
-
The protocol describes the lentivirus-based expression of high amounts of soluble and membrane-bound tagged proteins of interest (POI) in human suspension cells. This is combined with rapid nanobody-based purification of the POI and its complexes for downstream structural analysis and functional assays.
-
The protocol provides guidelines for the isolation of a POI in its tagged (TagON) and scarless untagged (TagOFF) forms using an engineered SUMO protease (SENPEuB).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The lentiviral transfer plasmids and bacterial expression plasmids described in this study are available from Addgene. Addgene IDs of all plasmids are listed in Table 1. Source data are provided with this paper.
References
Elegheert, J. et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat. Protoc. 13, 2991–3017 (2018).
Pleiner, T. et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science 369, 433–436 (2020).
Götzke, H. et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10, 4403 (2019).
Vera Rodriguez, A., Frey, S. & Görlich, D. Engineered SUMO/protease system identifies Pdr6 as a bidirectional nuclear transport receptor. J. Cell Biol. 218, 2006–2020 (2019).
Huh, W.-K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Hayashi, A. et al. Localization of gene products using a chromosomally tagged GFP‐fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells 14, 217–225 (2009).
Sarov, M. et al. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150, 855–866 (2012).
Sarov, M. et al. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, e12068 (2016).
Poser, I. et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 (2008).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Pleiner, T. et al. Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. eLife 4, e11349 (2015).
Aksu, M. et al. Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J. Cell Biol. 217, 2329–2340 (2018).
Fridy, P. C. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014).
De Genst, E. J. et al. Structure and properties of a complex of α-synuclein and a single-domain camelid antibody. J. Mol. Biol. 402, 326–343 (2010).
Braun, M. B. et al. Peptides in headlock–a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci. Rep. 6, 19211 (2016).
Virant, D. et al. A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat. Commun. 9, 930 (2018).
Strokappe, N. M. et al. Super potent bispecific llama VHH antibodies neutralize HIV via a combination of gp41 and gp120 epitopes. Antibodies 8, 38 (2019).
Traenkle, B., Segan, S., Fagbadebo, F. O., Kaiser, P. D. & Rothbauer, U. A novel epitope tagging system to visualize and monitor antigens in live cells with chromobodies. Sci. Rep. 10, 14267 (2020).
Xu, J. et al. Protein visualization and manipulation in Drosophila through the use of epitope tags recognized by nanobodies. eLife 11, e74326 (2022).
Lindborg, M. et al. High-affinity binding to staphylococcal protein A by an engineered dimeric Affibody molecule. Protein Eng. Des. Sel. 26, 635–644 (2013).
Vassylyeva, M. N. et al. Efficient, ultra-high-affinity chromatography in a one-step purification of complex proteins. Proc. Natl Acad. Sci. USA 114, E5138–E5147 (2017).
Chen, J. et al. Structure of an endogenous mycobacterial MCE lipid transporter. Nature, (2023).
Juszkiewicz, S. & Hegde, R. S. Quality control of orphaned proteins. Mol. Cell 71, 443–457 (2018).
Pleiner, T. et al. WNK1 is an assembly factor for the human ER membrane protein complex. Mol. Cell 81, 2693–2704.e12 (2021).
Guna, A. et al. MTCH2 is a mitochondrial outer membrane protein insertase. Science 378, 317–322 (2022).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Schmidt, T. G. et al. Development of the Twin-Strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif. 92, 54–61 (2013).
Qin, J. Y. et al. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE 5, e10611 (2010).
Zufferey, R., Donello, J. E., Trono, D. & Hope, T. J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73, 2886–2892 (1999).
de Felipe, P. et al. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 24, 68–75 (2006).
McShane, E. et al. Kinetic analysis of protein stability reveals age-dependent degradation. Cell 167, 803–815.e21 (2016).
Cronin, J., Zhang, X. Y. & Reiser, J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5, 387–398 (2005).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Chaudhary, S., Pak, J. E., Gruswitz, F., Sharma, V. & Stroud, R. M. Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nat. Protoc. 7, 453–466 (2012).
O’Gorman, S., Fox, D. T. & Wahl, G. M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251, 1351–1355 (1991).
Leonetti, M. D., Sekine, S., Kamiyama, D., Weissman, J. S. & Huang, B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl Acad. Sci. USA 113, E3501–E3508 (2016).
Koch, B. et al. Generation and validation of homozygous fluorescent knock-in cells using CRISPR–Cas9 genome editing. Nat. Protoc. 13, 1465–1487 (2018).
Cho, N. H. et al. OpenCell: endogenous tagging for the cartography of human cellular organization. Science 375, eabi6983 (2022).
Nilsen, T. W. Preparation of nuclear extracts from HeLa cells. Cold Spring Harb. Protoc. 2013, 579–583 (2013).
Seddon, A. M., Curnow, P. & Booth, P. J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta 1666, 105–117 (2004).
Lee, S. C. et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 11, 1149–1162 (2016).
Rothbauer, U. et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889 (2006).
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 (2010).
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Schatz, P. J. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology 11, 1138–1143 (1993).
Beckett, D., Kovaleva, E. & Schatz, P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 (1999).
Fairhead, M. & Howarth, M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 1266, 171–184 (2015).
Frey, S. & Görlich, D. The Xenopus laevis Atg4B protease: insights into substrate recognition and application for tag removal from proteins expressed in pro- and eukaryotic hosts. PLoS ONE 10, e0125099 (2015).
Liu, L., Spurrier, J., Butt, T. R. & Strickler, J. E. Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions. Protein Expr. Purif. 62, 21–28 (2008).
Voorhees, R. M. & Hegde, R. S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 4, e07975 (2015).
Frey, S. & Görlich, D. A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A 1337, 95–105 (2014).
Pleiner, T., Bates, M. & Görlich, D. A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J. Cell Biol. 217, 1143–1154 (2018).
Kubala, M. H., Kovtun, O., Alexandrov, K. & Collins, B. M. Structural and thermodynamic analysis of the GFP: GFP–nanobody complex. Protein Sci. 19, 2389–2401 (2010).
Acknowledgements
We thank P. Bjorkman for access to her laboratory’s cell sorter, as well as the Caltech Flow Cytometry facility. This work was supported by the Heritage Medical Research Institute (R.M.V.), the National Institutes of Health’s National Institute Of General Medical Sciences DP2GM137412 (R.M.V.), the Deutsche Forschungsgemeinschaft (T.P.) and the Tianqiao and Chrissy Chen Institute (T.P. and M.H.).
Author information
Authors and Affiliations
Contributions
T.A.S., R.M.V. and T.P. conceived and designed this study. T.A.S., G.P.T., M.H., S.W., V.N.N., C.D. and T.P. carried out the experiments and interpreted data. T.A.S., R.M.V. and T.P. wrote the protocol, and all authors provided feedback on its final version.
Corresponding authors
Ethics declarations
Competing interests
R.M.V. and G.P.T. are consultants for Gates Biosciences, and R.M.V. is an equity holder.
Peer review
Peer review information
Nature Protocols thanks Eric Gouaux and Jan Steyaert for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Pleiner, T. et al. eLife 4, e11349 (2015): https://doi.org/10.7554/eLife.11349
Pleiner, T. et al. Science 369, 433–436 (2020): https://doi.org/10.1126/science.abb5008
Guna, A. et al. Science 378, 317–322 (2022): https://doi.org/10.1126/science.add1856
Extended data
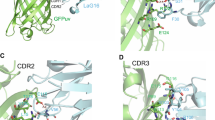
Extended Data Fig. 1 Overview of anti-GFP nanobody compatible fluorescent protein variants.
(a) Crystal structure of GFP bound to anti-GFP nanobody (Nb) (PDB ID: 3K1K)43 with GFP shown in green with cartoon rendering, anti-GFP Nb shown in blue with surface rendering, and specific residues on GFP that make contact with the anti-GFP Nb shown in stick rendering. (b) Front-view of the anti-GFP nanobody binding surface of GFP with participating residues shown in stick rendering. Residues mutated in other fluorescent protein variants colored in salmon. (c) Multiple sequence alignment of various fluorescent protein variants. Columns corresponding to residues contacted by the anti-GFP Nb are highlighted in yellow, and any mutations to these positions are highlighted in red. Mutation of I146N was previously shown to restore anti-GFP Nb binding in CFP variants53.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Fig. 1 and Data 1.
Supplementary Data 2
E. coli protein expression data sheet.
Source data
Source Data Fig. 3
Uncropped SDS–PAGE gels.
Source Data Fig. 4
Uncropped SDS–PAGE gels.
Source Data Fig. 5
Uncropped SDS–PAGE gels and western blots.
Source Data Fig. 6
Uncropped SDS–PAGE gel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Stevens, T.A., Tomaleri, G.P., Hazu, M. et al. A nanobody-based strategy for rapid and scalable purification of human protein complexes. Nat Protoc 19, 127–158 (2024). https://doi.org/10.1038/s41596-023-00904-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00904-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.