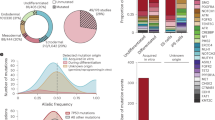
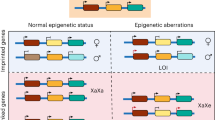
Abstract
Human pluripotent stem cells (hPSCs) are known to acquire genetic aberrations during in vitro propagation. In addition to recurrent chromosomal aberrations, it has recently been shown that these cells also gain point mutations in cancer-related genes, predominantly in TP53. The need for routine quality control of hPSCs is critical for both basic research and clinical applications. Here we discuss the relevance of detecting mutations for various hPSCs applications, and present a detailed protocol to identify cancer-related point mutations using data from RNA sequencing, an assay commonly performed during the growth and differentiation of hPSCs. In this protocol, we describe how to process and align the sequencing data, analyze it and conservatively interpret the results in order to generate an accurate estimation of mutations in tumor-related genes. This pipeline is designed to work in high throughput and is available as a software container at https://github.com/elyadlezmi/RNA2CM. The protocol requires minimal command-line skills and can be carried out in 1–2 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The RNA-seq sample used as an example in this protocol can be retrieved from the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) under the accession number SRR3090631.
Code availabilty
All the code used in this protocol is available at https://github.com/elyadlezmi/RNA2CM39.
References
De Los Angeles, A. et al. Hallmarks of pluripotency. Nature 525, 469–478 (2015).
Tabar, V. & Studer, L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat. Rev. Genet. 15, 82–92 (2014).
Avior, Y., Sagi, I. & Benvenisty, N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 17, 170–182 (2016).
Shahbazi, M. N., Siggia, E. D. & Zernicka-Goetz, M. Self-organization of stem cells into embryos: a window on early mammalian development. Science 364, 948–951 (2019).
Weissbein, U., Benvenisty, N. & Ben-David, U. Genome maintenance in pluripotent stem cells. J. Cell Biol. 204, 153–163 (2014).
Bar, S. & Benvenisty, N. Epigenetic aberrations in human pluripotent stem cells. EMBO J. 38, 1–18 (2019).
Na, J., Baker, D., Zhang, J., Andrews, P. W. & Barbaric, I. Aneuploidy in pluripotent stem cells and implications for cancerous transformation. Protein Cell 5, 569–579 (2014).
Jo, H. Y. et al. Functional in vivo and in vitro effects of 20q11.21 genetic aberrations on hPSC differentiation. Sci. Rep. 10, 1–14 (2020).
Ben-David, U. & Benvenisty, N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 11, 268–277 (2011).
Ben-David, U. et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 5, 4825 (2014).
Simonson, O. E., Domogatskaya, A., Volchkov, P. & Rodin, S. The safety of human pluripotent stem cells in clinical treatment. Ann. Med. 47, 370–380 (2015).
Gore, A. et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67 (2011).
Merkle, F. T. et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233 (2017).
Avior, Y., Lezmi, E., Eggan, K. & Benvenisty, N. Cancer-related mutations identified in primed human pluripotent stem cells. Cell Stem Cell 28, 10–11 (2021).
Stirparo, G. G., Smith, A. & Guo, G. Cancer-related mutations are not enriched in naive human pluripotent stem cells. Cell Stem Cell 28, 164–169.e2 (2021).
Halliwell, J., Barbaric, I. & Andrews, P. W. Acquired genetic changes in human pluripotent stem cells: origins and consequences. Nat. Rev. Mol. Cell Biol. 21, 715–728 (2020).
Merkle, F. T. et al. Biological insights from the whole genome analysis of human embryonic stem cells. Preprint at bioRxiv https://doi.org/10.1101/2020.10.26.337352 (2020).
Trounson, A. & DeWitt, N. D. Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 17, 194–200 (2016).
Tate, J. G. et al. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 47, D941–D947 (2019).
Shihab, H. A. et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34, 57–65 (2013).
Sherry, S. T. et al. DbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Yizhak, K. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019).
Coudray, A., Battenhouse, A. M., Bucher, P. & Iyer, V. R. Detection and benchmarking of somatic mutations in cancer genomes using RNA-seq data. PeerJ 6, (2018).
Weissbein, U., Schachter, M., Egli, D. & Benvenisty, N. Analysis of chromosomal aberrations and recombination by allelic bias in RNA-Seq. Nat. Commun. 7, 12144 (2016).
Radenbaugh, A. J. et al. RADIA: RNA and DNA integrated analysis for somatic mutation detection. PLoS One 9, e111516 (2014).
DI Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Merkel, D. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 239, 2 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Danecek, P. & McCarthy, S. A. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics 33, 2037–2039 (2017).
Li, H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27, 718–719 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Brouard, J.-S., Schenkel, F., Marete, A. & Bissonnette, N. The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. J. Anim. Sci. Biotechnol. 10, 44 (2019).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018).
Kluin, R. J. C. et al. XenofilteR: computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinformatics 19, 366 (2018).
Collinson, A. et al. Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells article deletion of the Polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in Hu. Cell Rep. 17, 2700–2714 (2016).
Lezmi, E. Identification of cancer-related mutations in human pluripotent stem cells utilizing RNA-seq analysis. elyadlezmi/RNA2CM https://doi.org/10.5281/zenodo.4810015 (2021).
Acknowledgements
We thank S. Kinreich and A. Pagis for testing the pipeline and providing their constructive input and all members of The Azrieli Center for Stem Cells and Genetic Research for critical reading of the manuscript. This work was partially supported by the Israel Science Foundation (494/17), the Rosetrees Trust, and Azrieli Foundation. N.B. is the Herbert Cohn Chair in Cancer Research.
Author information
Authors and Affiliations
Contributions
E.L. and N.B. designed the analysis. E.L. developed the bioinformatic pipeline and wrote the manuscript with input from N.B., who supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Anna Esteve-Codina and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Merkle, F. et al. Nature 545, 229–233 (2017): https://doi.org/10.1038/nature22312
Avior, Y. et al. Cell Stem Cell 28, 10–11 (2021): https://doi.org/10.1016/j.stem.2020.11.013
Rights and permissions
About this article
Cite this article
Lezmi, E., Benvenisty, N. Identification of cancer-related mutations in human pluripotent stem cells using RNA-seq analysis. Nat Protoc 16, 4522–4537 (2021). https://doi.org/10.1038/s41596-021-00591-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00591-5
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.