Abstract
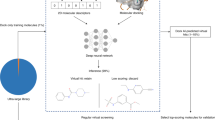
Structure-based virtual screening (SBVS) via docking has been used to discover active molecules for a range of therapeutic targets. Chemical and protein data sets that contain integrated bioactivity information have increased both in number and in size. Artificial intelligence and, more concretely, its machine-learning (ML) branch, including deep learning, have effectively exploited these data sets to build scoring functions (SFs) for SBVS against targets with an atomic-resolution 3D model (e.g., generated by X-ray crystallography or predicted by AlphaFold2). Often outperforming their generic and non-ML counterparts, target-specific ML-based SFs represent the state of the art for SBVS. Here, we present a comprehensive and user-friendly protocol to build and rigorously evaluate these new SFs for SBVS. This protocol is organized into four sections: (i) using a public benchmark of a given target to evaluate an existing generic SF; (ii) preparing experimental data for a target from public repositories; (iii) partitioning data into a training set and a test set for subsequent target-specific ML modeling; and (iv) generating and evaluating target-specific ML SFs by using the prepared training-test partitions. All necessary code and input/output data related to three example targets (acetylcholinesterase, HMG-CoA reductase, and peroxisome proliferator-activated receptor-α) are available at https://github.com/vktrannguyen/MLSF-protocol, can be run by using a single computer within 1 week and make use of easily accessible software/programs (e.g., Smina, CNN-Score, RF-Score-VS and DeepCoy) and web resources. Our aim is to provide practical guidance on how to augment training data to enhance SBVS performance, how to identify the most suitable supervised learning algorithm for a data set, and how to build an SF with the highest likelihood of discovering target-active molecules within a given compound library.
Key points
-
Scoring functions (SFs) can identify the compounds most likely to have a desired activity from their docked poses on an atomic-resolution structure of the considered macromolecular target. SFs built by using machine learning often outperform their classical counterparts.
-
This protocol describes how to use machine learning to build target-specific SFs and, importantly, how to assess how well they discriminate between actives and inactives of that target in chemically diverse libraries.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All input and output data involved in this protocol can be downloaded from https://github.com/vktrannguyen/MLSF-protocol.
References
Pereira, D. A. & Williams, J. A. Origin and evolution of high throughput screening. Br. J. Pharmacol. 152, 53–61 (2007).
Wang, Y., Cheng, T. & Bryant, S. H. PubChem BioAssay: a decade’s development toward open high-throughput screening data sharing. SLAS Discov. 22, 655–666 (2017).
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
Heifetz, A., Southey, M., Morao, I., Townsend-Nicholson, A. & Bodkin, M. J. Computational methods used in hit-to-lead and lead optimization stages of structure-based drug discovery. Methods Mol. Biol. 1705, 375–394 (2018).
Jorgensen, W. L. Efficient drug lead discovery and optimization. Acc. Chem. Res. 42, 724–733 (2009).
Gloriam, D. E. Bigger is better in virtual drug screens. Nature 566, 193–194 (2019).
Jia, C.-Y., Li, J.-Y., Hao, G.-F. & Yang, G.-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today 25, 248–258 (2020).
Göller, A. H. et al. Bayer’s in silico ADMET platform: a journey of machine learning over the past two decades. Drug Discov. Today 25, 1702–1709 (2020).
Grygorenko, O. O. et al. Generating multibillion chemical space of readily accessible screening compounds. iScience 23, 101681 (2020).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019).
Gorgulla, C. et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668 (2020).
Stein, R. M. et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020).
Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell 180, 688–702 (2020).
Gorgulla, C. et al. A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening. iScience 24, 102021 (2021).
Luttens, A. et al. Ultralarge virtual screening identifies SARS-CoV-2 main protease inhibitors with broad-spectrum activity against coronaviruses. J. Am. Chem. Soc. 144, 2905–2920 (2022).
Crunkhorn, S. Screening ultra-large virtual libraries. Nat. Rev. Drug Discov. 21, 95 (2022).
Fresnais, L. & Ballester, P. J. The impact of compound library size on the performance of scoring functions for structure-based virtual screening. Brief. Bioinform. 22, bbaa095 (2021).
Koes, D. R., Baumgartner, M. P. & Camacho, C. J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 53, 1893–1904 (2013).
Bender, B. J. et al. A practical guide to large-scale docking. Nat. Protoc. 16, 4799–4832 (2021).
Ain, Q. U., Aleksandrova, A., Roessler, F. D. & Ballester, P. J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdiscip. Rev. Comput. Mol. Sci. 5, 405–424 (2015).
Ballester, P. J. & Mitchell, J. B. O. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 26, 1169–1175 (2010).
Xiong, G.-L. et al. Improving structure-based virtual screening performance via learning from scoring function components. Brief. Bioinform. 22, bbaa094 (2021).
Li, H., Sze, K.-H., Lu, G. & Ballester, P. J. Machine-learning scoring functions for structure-based virtual screening. Wiley Interdiscip. Rev. Comput. Mol. Sci. 11, e1478 (2021).
Adeshina, Y. O., Deeds, E. J. & Karanicolas, J. Machine learning classification can reduce false positives in structure-based virtual screening. Proc. Natl Acad. Sci. USA 117, 18477–18488 (2020).
Nguyen, D. D. et al. Mathematical deep learning for pose and binding affinity prediction and ranking in D3R Grand Challenges. J. Comput. Aided Mol. Des. 33, 71–82 (2019).
Nguyen, D. D., Gao, K., Wang, M. & Wei, G. W. MathDL: mathematical deep learning for D3R Grand Challenge 4. J. Comput. Aided Mol. Des. 34, 131–147 (2020).
Li, H., Sze, K.-H., Lu, G. & Ballester, P. J. Machine-learning scoring functions for structure-based drug lead optimization. Wiley Interdiscip. Rev. Comput. Mol. Sci. 10, e1465 (2020).
Li, H. et al. Classical scoring functions for docking are unable to exploit large volumes of structural and interaction data. Bioinformatics 35, 3989–3995 (2019).
Meng, Z. & Xia, K. Persistent spectral–based machine learning (PerSpect ML) for protein-ligand binding affinity prediction. Sci. Adv. 7, eabc5329 (2021).
Shen, C. et al. From machine learning to deep learning: advances in scoring functions for protein–ligand docking. Wiley Interdiscip. Rev. Comput. Mol. Sci. 10, e1429 (2020).
Jiménez-Luna, J. et al. DeltaDelta neural networks for lead optimization of small molecule potency. Chem. Sci. 10, 10911–10918 (2019).
Sánchez-Cruz, N., Medina-Franco, J. L., Mestres, J. & Barril, X. Extended connectivity interaction features: improving binding affinity prediction through chemical description. Bioinformatics 37, 1376–1382 (2021).
Boyles, F., Deane, C. M. & Morris, G. M. Learning from docked ligands: ligand-based features rescue structure-based scoring functions when trained on docked poses. J. Chem. Inf. Model. 62, 5329–5341 (2022).
Li, H. et al. The impact of protein structure and sequence similarity on the accuracy of machine-learning scoring functions for binding affinity prediction. Biomolecules 8, 12 (2018).
Cang, Z., Mu, L. & Wei, G.-W. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput. Biol. 14, e1005929 (2018).
Jiang, P. et al. Molecular persistent spectral image (Mol-PSI) representation for machine learning models in drug design. Brief. Bioinform. 23, bbab527 (2022).
Wang, Z. et al. OnionNet-2: a convolutional neural network model for predicting protein-ligand binding affinity based on residue-atom contacting shells. Front. Chem. 9, 753002 (2021).
Karlov, D. S., Sosnin, S., Fedorov, M. V. & Popov, P. graphDelta: MPNN scoring function for the affinity prediction of protein-ligand complexes. ACS Omega 5, 5150–5159 (2020).
Tran-Nguyen, V. K. & Ballester, P. J. Beware of simple methods for structure-based virtual screening: the critical importance of broader comparisons. J. Chem. Inf. Model. 63, 1401–1405 (2023).
Wójcikowski, M., Ballester, P. J. & Siedlecki, P. Performance of machine-learning scoring functions in structure-based virtual screening. Sci. Rep. 7, 46710 (2017).
Li, H., Leung, K.-S., Wong, M.-H. & Ballester, P. J. Correcting the impact of docking pose generation error on binding affinity prediction. BMC Bioinforma. 17, 308 (2016).
Coleman, R. G., Carchia, M., Sterling, T., Irwin, J. J. & Shoichet, B. K. Ligand pose and orientational sampling in molecular docking. PLoS One 8, e75992 (2013).
Ragoza, M., Hochuli, J., Idrobo, E., Sunseri, J. & Koes, D. R. Protein–ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 (2017).
Imrie, F., Bradley, A. R., van der Schaar, M. & Deane, C. M. Protein family-specific models using deep neural networks and transfer learning improve virtual screening and highlight the need for more data. J. Chem. Inf. Model. 58, 2319–2330 (2018).
Ghislat, G., Rahman, T. & Ballester, P. J. Recent progress on the prospective application of machine learning to structure-based virtual screening. Curr. Opin. Chem. Biol. 65, 28–34 (2021).
Durrant, J. D. et al. Neural-network scoring functions identify structurally novel estrogen-receptor ligands. J. Chem. Inf. Model. 55, 1953–1961 (2015).
Sun, H. et al. Constructing and validating high-performance MIEC-SVM models in virtual screening for kinases: a better way for actives discovery. Sci. Rep. 6, 24817 (2016).
Stecula, A., Hussain, M. S. & Viola, R. E. Discovery of novel inhibitors of a critical brain enzyme using a homology model and a deep convolutional neural network. J. Med. Chem. 63, 8867–8875 (2020).
Yasuo, N. & Sekijima, M. An improved method of structure-based virtual screening via interaction-energy-based learning. J. Chem. Inf. Model. 59, 1050–1061 (2019).
Wijewardhane, P. R., Jethava, K. P., Fine, J. A. & Chopra, G. Combined molecular graph neural network and structural docking selects potent programmable cell death protein 1/programmable death-ligand 1 (PD-1/PD-L1) small molecule inhibitors. Preprint at https://chemrxiv.org/engage/chemrxiv/article-details/60c74991bb8c1a15b13dae70 (2020).
Doman, T. N. et al. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J. Med. Chem. 45, 2213–2221 (2002).
Shoichet, B. K., Stroud, R. M., Santi, D. V., Kuntz, I. D. & Perry, K. M. Structure-based discovery of inhibitors of thymidylate synthase. Science 259, 1445–1450 (1993).
Gentile, F. et al. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat. Protoc. 17, 672–697 (2022).
Ashtawy, H. M. & Mahapatra, N. R. Machine-learning scoring functions for identifying native poses of ligands docked to known and novel proteins. BMC Bioinforma. 16 (Suppl 6), S3 (2015).
Bauer, M. R., Ibrahim, T. M., Vogel, S. M. & Boeckler, F. M. Evaluation and optimization of virtual screening workflows with DEKOIS 2.0—a public library of challenging docking benchmark sets. J. Chem. Inf. Model. 53, 1447–1462 (2013).
Marcou, G. & Rognan, D. Optimizing fragment and scaffold docking by use of molecular interaction fingerprints. J. Chem. Inf. Model. 47, 195–207 (2007).
Zhan, W. et al. Integrating docking scores, interaction profiles and molecular descriptors to improve the accuracy of molecular docking: toward the discovery of novel Akt1 inhibitors. Eur. J. Med. Chem. 75, 11–20 (2014).
Mir, S. et al. PDBe: towards reusable data delivery infrastructure at protein data bank in Europe. Nucleic Acids Res. 46, D486–D492 (2018).
Harrison, C. Homology model allows effective virtual screening. Nat. Rev. Drug Discov. 10, 816 (2011).
Huang, D. et al. On the value of homology models for virtual screening: discovering hCXCR3 antagonists by pharmacophore-based and structure-based approaches. J. Chem. Inf. Model. 52, 1356–1366 (2012).
Messaoudi, A., Belguith, H. & Hamida, J. B. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor. Biol. Med. Model. 10, 22 (2013).
Chen, X.-R. et al. Homology modeling and virtual screening to discover potent inhibitors targeting the imidazole glycerophosphate dehydratase protein in Staphylococcus xylosus. Front. Chem. 5, 98 (2017).
Leffler, A. E. et al. Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models. Proc. Natl Acad. Sci. USA 114, E8100–E8109 (2017).
Jaiteh, M., Rodríguez-Espigares, I., Selent, J. & Carlsson, J. Performance of virtual screening against GPCR homology models: impact of template selection and treatment of binding site plasticity. PloS Comput. Biol. 16, e1007680 (2020).
Panda, S. K., Saxena, S. & Guruprasad, L. Homology modeling, docking and structure-based virtual screening for new inhibitor identification of Klebsiella pneumoniae heptosyltransferase-III. J. Biomol. Struct. Dyn. 38, 1887–1902 (2020).
Kopp, J. & Schwede, T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 32, D230–D234 (2004).
Bienert, S. et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 45, D313–D319 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Callaway, E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 588, 203–204 (2020).
Callaway, E. What’s next for AlphaFold and the AI protein-folding revolution. Nature 604, 234–238 (2022).
Ren, F. et al. AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chem. Sci. 14, 1443–1452 (2023).
Wong, F. et al. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 18, e11081 (2022).
Ballester, P. J. Selecting machine-learning scoring functions for structure-based virtual screening. Drug Discov. Today Technol. 32–33, 81–87 (2020).
Xiong, G. et al. Featurization strategies for protein–ligand interactions and their applications in scoring function development. Wiley Interdiscip. Rev. Comput. Mol. Sci. 12, e1567 (2021).
Huang, N., Shoichet, B. K. & Irwin, J. J. Benchmarking sets for molecular docking. J. Med. Chem. 49, 6789–6801 (2006).
Vogel, S. M., Bauer, M. R. & Boeckler, F. M. DEKOIS: demanding evaluation kits for objective in silico screening—a versatile tool for benchmarking docking programs and scoring functions. J. Chem. Inf. Model. 51, 2650–2665 (2011).
Mysinger, M. M., Carchia, M., Irwin, J. J. & Shoichet, B. K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594 (2012).
Rohrer, S. G. & Baumann, K. Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J. Chem. Inf. Model. 49, 169–184 (2009).
Tran-Nguyen, V. K., Jacquemard, C. & Rognan, D. LIT-PCBA: an unbiased data set for machine learning and virtual screening. J. Chem. Inf. Model. 60, 4263–4273 (2020).
Wallach, I. & Heifets, A. Most ligand-based classification benchmarks reward memorization rather than generalization. J. Chem. Inf. Model. 58, 916–932 (2018).
Tran-Nguyen, V. K. & Rognan, D. Benchmarking data sets from PubChem BioAssay data: current scenario and room for improvement. Int. J. Mol. Sci. 21, 4380 (2020).
Lagarde, N., Zagury, J.-F. & Montes, M. Benchmarking data sets for the evaluation of virtual ligand screening methods: review and perspectives. J. Chem. Inf. Model. 55, 1297–1307 (2015).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 (2011).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Dos Santos, R. N., Ferreira, L. G. & Andricopulo, A. D. Practices in molecular docking and structure-based virtual screening. Methods Mol. Biol. 1762, 31–50 (2018).
Da Silva, F., Desaphy, J. & Rognan, D. IChem: a versatile toolkit for detecting, comparing, and predicting protein-ligand interactions. ChemMedChem 13, 507–510 (2018).
Tran-Nguyen, V. K., Da Silva, F., Bret, G. & Rognan, D. All in one: cavity detection, druggability estimate, cavity-based pharmacophore perception, and virtual screening. J. Chem. Inf. Model. 59, 573–585 (2019).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Tran-Nguyen, V. K., Simeon, S., Junaid, M. & Ballester, P. J. Structure-based virtual screening for PDL1 dimerizers: evaluating generic scoring functions. Curr. Res. Struct. Biol. 4, 206–210 (2022).
Eriksson, L. et al. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ. Health Perspect. 111, 1361–1375 (2003).
Sahigara, F. et al. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 17, 4791–4810 (2012).
Carrio, P., Pinto, M., Ecker, G., Sanz, F. & Pastor, M. Applicability domain analysis (ADAN): a robust method for assessing the reliability of drug property predictions. J. Chem. Inf. Model. 54, 1500–1511 (2014).
Sahlin, U., Jeliazkova, N. & Öberg, T. Applicability domain dependent predictive uncertainty in QSAR regressions. Mol. Inform. 33, 26–35 (2014).
Kaneko, H. & Funatsu, K. Applicability domain based on ensemble learning in classification and regression analyses. J. Chem. Inf. Model. 54, 2469–2482 (2014).
Ballester, P. J. & Mitchell, J. B. O. Comments on “Leave-cluster-out cross-validation is appropriate for scoring functions derived from diverse protein data sets”: significance for the validation of scoring functions. J. Chem. Inf. Model. 51, 1739–1741 (2011).
Tran-Nguyen, V. K., Bret, G. & Rognan, D. True accuracy of fast scoring functions to predict high-throughput screening data from docking poses: the simpler the better. J. Chem. Inf. Model. 61, 2788–2797 (2021).
Stepniewska-Dziubinska, M. M., Zielenkiewicz, P. & Siedlecki, P. Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 34, 3666–3674 (2018).
Wang, C. & Zhang, Y. Improving scoring-docking-screening powers of protein-ligand scoring functions using random forest. J. Comput. Chem. 38, 169–177 (2017).
Shen, C. et al. Accuracy or novelty: what can we gain from target-specific machine-learning-based scoring functions in virtual screening? Brief. Bioinform. 22, bbaa410 (2021).
McNutt, A. T. et al. GNINA 1.0: molecular docking with deep learning. J. Cheminform. 13, 43 (2021).
Saito, T. & Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PloS One 10, e0118432 (2015).
Liu, S. et al. Practical model selection for prospective virtual screening. J. Chem. Inf. Model. 59, 282–293 (2019).
Mendez, D. et al. ChEMBL: toward direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2019).
Papadatos, G. et al. SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 44, D1220–D1228 (2016).
Sunghwan, K. et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 49, D1388–D1395 (2021).
McCloskey, K. et al. Machine learning on DNA-encoded libraries: a new paradigm for hit finding. J. Med. Chem. 63, 8857–8866 (2020).
Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Gilberg, E., Jasial, S., Stumpfe, D., Dimova, D. & Bajorath, J. Highly promiscuous small molecules from biological screening assays include many pan-assay interference compounds but also candidates for polypharmacology. J. Med. Chem. 59, 10285–10290 (2016).
Baell, J. B. Feeling nature’s PAINS: natural products, natural product drugs, and pan assay interference compounds (PAINS). J. Nat. Prod. 79, 616–628 (2016).
Capuzzi, S. J., Muratov, E. N. & Tropsha, A. Phantom PAINS: problems with the utility of alerts for Pan-Assay INterference CompoundS. J. Chem. Inf. Model. 57, 417–427 (2017).
Kenny, P. W. Comment on the ecstasy and agony of assay interference compounds. J. Chem. Inf. Model. 57, 2640–2645 (2017).
Baell, J. B. & Nissink, J. W. Seven year itch: pan-assay interference compounds (PAINS) in 2017—utility and limitations. ACS Chem. Biol. 13, 36–44 (2018).
Stork, C., Chen, Y., Sicho, M. & Kirchmair, J. Hit Dexter 2.0: machine-learning models for the prediction of frequent hitters. J. Chem. Inf. Model. 59, 1030–1043 (2019).
Stork, C. et al. NERDD: a web portal providing access to in silico tools for drug discovery. Bioinformatics 36, 1291–1292 (2020).
Pearl, L. H. Review: the HSP90 molecular chaperone-an enigmatic ATPase. Biopolymers 105, 594–607 (2016).
Sgobba, M., Forestiero, R., Degliesposti, G. & Rastelli, G. Exploring the binding site of C-terminal hsp90 inhibitors. J. Chem. Inf. Model. 50, 1522–1528 (2010).
Halgren, T. A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 49, 377–389 (2009).
Molecular Operating Environment (MOE), 2020.09. Chemical Computing Group https://www.chemcomp.com/Products.htm (2022).
Smyth, M. S. & Martin, J. H. J. x Ray crystallography. Mol. Pathol. 53, 8–14 (2000).
Wüthrich, K. Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 265, 22059–22062 (1990).
Purslow, J. A., Khatiwada, B., Bayro, M. J. & Venditti, V. NMR methods for structural characterization of protein-protein complexes. Front. Mol. Biosci. 7, 9 (2020).
Fowler, N. J., Sljoka, A. & Williamson, M. P. A method for validating the accuracy of NMR protein structures. Nat. Commun. 11, 6321 (2020).
Hu, Y. et al. NMR-based methods for protein analysis. Anal. Chem. 93, 1866–1879 (2021).
Callaway, E. Revolutionary cryo-EM is taking over structural biology. Nature 578, 201 (2020).
Wu, X. & Rapoport, T. A. Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies). Proc. Natl Acad. Sci. USA 118, e2115001118 (2021).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Oleinikovas, V., Saladino, G., Cossins, B. P. & Gervasio, F. L. Understanding cryptic pocket formation in protein targets by enhanced sampling simulations. J. Am. Chem. Soc. 138, 14257–14263 (2016).
Vajda, S., Beglov, D., Wakefield, A. E., Egbert, M. & Whitty, A. Cryptic binding sites on proteins: definition, detection, and druggability. Curr. Opin. Chem. Biol. 44, 1–8 (2018).
Bekker, G. J., Fukuda, I., Higo, J., Fukunishi, Y. & Kamiya, N. Cryptic-site binding mechanism of medium-sized Bcl-xL inhibiting compounds elucidated by McMD-based dynamic docking simulations. Sci. Rep. 11, 5046 (2021).
Zhu, J., Hoop, C. L., Case, D. A. & Baum, J. Cryptic binding sites become accessible through surface reconstruction of the type I collagen fibril. Sci. Rep. 8, 16646 (2018).
Posner, B. A., Xi, H. & Mills, J. E. Enhanced HTS hit selection via a local hit rate analysis. J. Chem. Inf. Model. 49, 2202–2210 (2009).
Stein, R. M. et al. Property-unmatched decoys in docking benchmarks. J. Chem. Inf. Model. 61, 699–714 (2021).
Imrie, F., Bradley, A. R. & Deane, C. M. Generating property-matched decoy molecules using deep learning. Bioinformatics 37, 2134–2141 (2021).
Irwin, J. J., Sterling, T., Mysinger, M. M., Bolstad, E. S. & Coleman, R. G. ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 52, 1757–1768 (2012).
Réau, M., Langenfeld, F., Zagury, J.-F., Lagarde, N. & Montes, M. Decoys selection in benchmarking datasets: overview and perspectives. Front. Pharmacol. 9, 11 (2018).
Moriwaki, H., Tian, Y.-S., Kawashita, N. & Takagi, T. Mordred: a molecular descriptor calculator. J. Cheminform. 10, 4 (2018).
Barillari, C., Taylor, J., Viner, R. & Essex, J. W. Classification of water molecules in protein binding sites. J. Am. Chem. Soc. 129, 2577–2587 (2007).
Liu, T., Lin, Y., Wen, X., Jorissen, R. N. & Gilson, M. K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 35, D198–D201 (2007).
Hernández-Hernández, S. & Ballester, P. J. On the best way to cluster NCI-60 molecules. Biomolecules 13, 498 (2023).
Butina, D. Unsupervised data base clustering based on Daylight’s fingerprint and Tanimoto similarity: a fast and automated way to cluster small and large data sets. J. Chem. Inf. Comput. Sci. 39, 747–750 (1999).
Gómez-Sacristán, P. et al. Structure-based virtual screening for PDL1 dimerizers is boosted by inactive-enriched machine-learning models exploiting patent data. Zenodo https://zenodo.org/record/6226320/export/dcite4 (2023).
Radifar, M., Yuniarti, N. & Istyastono, E. P. PyPLIF: Python-based protein-ligand interaction fingerprinting. Bioinformation 9, 325–328 (2013).
Chupakhin, V., Marcou, G., Gaspar, H. & Varnek, A. Simple ligand–receptor interaction descriptor (SILIRID) for alignment-free binding site comparison. Comput. Struct. Biotechnol. J. 10, 33–37 (2014).
Da, C. & Kireev, D. Structural protein–ligand interaction fingerprints (SPLIF) for structure-based virtual screening: method and benchmark study. J. Chem. Inf. Model. 54, 2555–2561 (2014).
Ballester, P. J., Schreyer, A. & Blundell, T. L. Does a more precise chemical description of protein-ligand complexes lead to more accurate prediction of binding affinity? J. Chem. Inf. Model. 54, 944–955 (2014).
Li, H., Leung, K.-S., Wong, M.-H. & Ballester, P. J. Improving AutoDock Vina using Random Forest: the growing accuracy of binding affinity prediction by the effective exploitation of larger data sets. Mol. Inform. 34, 115–126 (2015).
Wójcikowski, M., Kukiełka, M., Stepniewska-Dziubinska, M. M. & Siedlecki, P. Development of a protein-ligand extended connectivity (PLEC) fingerprint and its application for binding affinity predictions. Bioinformatics 35, 1334–1341 (2019).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Ballester, P. J. et al. Hierarchical virtual screening for the discovery of new molecular scaffolds in antibacterial hit identification. J. R. Soc. Interface 9, 3196–3207 (2012).
Li, L. et al. Target-specific support vector machine scoring in structure-based virtual screening: computational validation, in vitro testing in kinases, and effects on lung cancer cell proliferation. J. Chem. Inf. Model. 51, 755–759 (2011).
Durrant, J. D. & McCammon, J. A. NNScore: a neural-network-based scoring function for the characterization of protein−ligand complexes. J. Chem. Inf. Model. 50, 1865–1871 (2010).
Durrant, J. D. & McCammon, J. A. NNScore 2.0: a neural-network receptor–ligand scoring function. J. Chem. Inf. Model. 51, 2897–2903 (2011).
Wang, D. et al. Improving the virtual screening ability of target-specific scoring functions using deep learning methods. Front. Pharmacol. 10, 924 (2019).
Ashtawy, H. M. & Mahapatra, N. R. Task-specific scoring functions for predicting ligand binding poses and affinity and for screening enrichment. J. Chem. Inf. Model. 58, 119–133 (2018).
Turner, R. et al. Bayesian optimization is superior to random search for machine learning hyperparameter tuning: analysis of the Black-Box Optimization Challenge 2020. Proc. Mach. Learn. Res. 133, 3–26 (2021).
Cowen-Rivers, A. I. et al. HEBO: pushing the limits of sample-efficient hyperparameter optimisation. J. Artif. Intell. Res. 74, 1269–1349 (2022).
Akiba, T., Sano, S., Yanase, T., Ohta, T. & Koyama, M. Optuna: a next-generation hyperparameter optimization framework. in The 25th ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD ’19), August 4–8, 2019, Anchorage, AK, USA. https://doi.org/10.1145/3292500.3330701 (2019).
Case, D. A. et al. The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Götz, A. W. et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555 (2012).
Berendsen, H. J. C., van der Spoel, D. & van Drunen, R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995).
Makarewicz, T. & Kaźmierkiewicz, R. Molecular dynamics simulation by GROMACS using GUI plugin for PyMOL. J. Chem. Inf. Model. 53, 1229–1234 (2013).
van Dijk, M., Wassenaar, T. A. & Bonvin, A. M. J. J. A flexible, grid-enabled web portal for GROMACS molecular dynamics simulations. J. Chem. Theory Comput. 8, 3463–3472 (2012).
Bietz, S., Urbaczek, S., Schulz, B. & Rarey, M. Protoss: a holistic approach to predict tautomers and protonation states in protein-ligand complexes. J. Cheminform. 6, 12 (2014).
Sunseri, J. & Koes, D. R. Virtual screening with Gnina 1.0. Molecules 26, 7369 (2021).
Acknowledgements
The authors acknowledge support from the French Association for Cancer Research (ARC), the Indo-French Centre for the Promotion of Advanced Research (CEFIPRA), the French National Research Agency (ANR), The Wolfson Foundation and the Royal Society for a Royal Society Wolfson Fellowship awarded to P.J.B.
Author information
Authors and Affiliations
Contributions
P.J.B. designed the protocol. P.J.B. developed the protocol with the assistance of V.-K.T.-N. V.-K.T.-N. generated the code repository by reusing code from previous publications and from S.S. V.-K.T.-N. and M.J. tested the protocol. P.J.B. and V.-K.T.-N. analyzed the results and wrote the paper with feedback from M.J. and S.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Brian Shoichet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Fresnais, L. & Ballester, P. J. Brief. Bioinform. 22, bbaa095 (2021): https://doi.org/10.1093/bib/bbaa095
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, Tables 1–5 and Discussion
Supplementary Data
PDB X-Ray Structure Validation Report
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tran-Nguyen, VK., Junaid, M., Simeon, S. et al. A practical guide to machine-learning scoring for structure-based virtual screening. Nat Protoc 18, 3460–3511 (2023). https://doi.org/10.1038/s41596-023-00885-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00885-w
This article is cited by
-
Comprehensive machine learning boosts structure-based virtual screening for PARP1 inhibitors
Journal of Cheminformatics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.