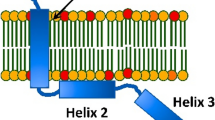
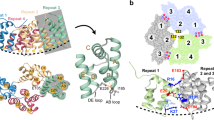
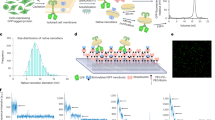
Abstract
The transmembrane (TM) anchors of cell surface proteins have been one of the ‘blind spots’ in structural biology because they are generally very hydrophobic, sometimes dynamic, and thus difficult targets for structural characterization. A plethora of examples show these membrane anchors are not merely anchors but can multimerize specifically to activate signaling receptors on the cell surface or to stabilize envelope proteins in viruses. Through a series of studies of the TM domains (TMDs) of immune receptors and viral membrane proteins, we have established a robust protocol for determining atomic-resolution structures of TM oligomers by NMR in bicelles that closely mimic a lipid bilayer. Our protocol overcomes hurdles typically encountered by structural biology techniques such as X-ray crystallography and cryo-electron microscopy (cryo-EM) when studying small TMDs. Here, we provide the details of the protocol, covering five major technical aspects: (i) a general method for producing isotopically labeled TM or membrane-proximal (MP) protein fragments that involves expression of the protein (which is fused to TrpLE) into inclusion bodies and releasing the target protein by cyanogen bromide (CNBr) cleavage; (ii) determination of the oligomeric state of TMDs in bicelles; (iii) detection of intermolecular contacts using nuclear Overhauser effect (NOE) experiments; (iv) structure determination; and (v) paramagnetic probe titration (PPT) to characterize the membrane partition of the TM oligomers. This protocol is broadly applicable for filling structural gaps of many type I/II membrane proteins. The procedures may take 3–6 months to complete, depending on the complexity and stability of the protein sample.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Code availability
The code and instructions for the ExSSO program are freely accessible from the website: http://www.csbio.sjtu.edu.cn/bioinf/ExSSO/. The software is also provided as Supplementary Software 1 and 2.
References
Call, M. E., Wucherpfennig, K. W. & Chou, J. J. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat. Immunol. 11, 1023–1029 (2010).
Call, M. E. & Wucherpfennig, K. W. Common themes in the assembly and architecture of activating immune receptors. Nat. Rev. Immunol. 7, 841–850 (2007).
Endres, N. F. et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152, 543–556 (2013).
Arkhipov, A. et al. Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569 (2013).
Fu, Q. et al. Structural basis and functional role of intramembrane trimerization of the Fas/CD95 death receptor. Mol. Cell 61, 602–613 (2016).
MacKenzie, K. R., Prestegard, J. H. & Engelman, D. M. A transmembrane helix dimer: structure and implications. Science 276, 131–133 (1997).
Trenker, R., Call, M. E. & Call, M. J. Crystal structure of the glycophorin a transmembrane dimer in lipidic cubic phase. J. Am. Chem. Soc. 137, 15676–15679 (2015).
Call, M. E. et al. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 127, 355–368 (2006).
Bocharov, E. V. et al. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 283, 6950–6956 (2008).
Lau, T. L., Kim, C., Ginsberg, M. H. & Ulmer, T. S. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28, 1351–1361 (2009).
Barrett, P. J. et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 336, 1168–1171 (2012).
Dev, J. et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science 353, 172–175 (2016).
Chen, W. et al. Familial Alzheimer’s mutations within APPTM increase Abeta42 production by enhancing accessibility of epsilon-cleavage site. Nat. Commun. 5, 3037 (2014).
Lee, J. et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. USA 114, E7987–E7996 (2017).
Klammt, C. et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat. Methods 9, 834 (2012).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Gasteiger, E. et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Glover, K. J. et al. Structural evaluation of phospholipid bicelles for solution-state studies of membrane-associated biomolecules. Biophys. J. 81, 2163–2171 (2001).
Piai, A., Fu, Q., Dev, J. & Chou, J. J. Optimal bicelle size q for solution NMR studies of the protein transmembrane partition. Chemistry 23, 1361–1367 (2017).
Sanders, C. R., Hare, B. J., Howard, K. P. & Prestegard, J. H. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog. Nucl. Magn. Reson. Spectrosc. 26, 421–444 (1994).
Caldwell, T. A. et al. Low-q bicelles are mixed micelles. J. Phys. Chem. Lett. 9, 4469–4473 (2018).
Piai, A., Dev, J., Fu, Q. & Chou, J. J. Stability and water accessibility of the trimeric membrane anchors of the HIV-1 envelope spikes. J. Am. Chem. Soc. 139, 18432–18435 (2017).
Chen, W. et al. The unusual transmembrane partition of the hexameric channel of the hepatitis C virus. Structure 26, 627–634.e4 (2018).
Fu, Q. et al. Structure of the membrane proximal external region of HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 115, E8892–E8899 (2018).
Knoblich, K. et al. Transmembrane complexes of DAP12 crystallized in lipid membranes provide insights into control of oligomerization in immunoreceptor assembly. Cell Rep. 11, 1184–1192 (2015).
Hofer, N., Aragao, D. & Caffrey, M. Crystallizing transmembrane peptides in lipidic mesophases. Biophys. J. 99, L23–L25 (2010).
Thomaston, J. L. & DeGrado, W. F. Crystal structure of the drug-resistant S31N influenza M2 proton channel. Protein Sci. 25, 1551–1554 (2016).
Cooper, R. S., Georgieva, E. R., Borbat, P. P., Freed, J. H. & Heldwein, E. E. Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat. Struct. Mol. Biol. 25, 416–424 (2018).
Barbet-Massin, E. et al. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J. Am. Chem. Soc. 136, 12489–12497 (2014).
Andreas, L. B. et al. Structure and mechanism of the influenza A M218-60 dimer of dimers. J. Am. Chem. Soc. 137, 14877–14886 (2015).
van Dam, L., Karlsson, G. & Edwards, K. Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochim. Biophys. Acta 1664, 241–256 (2004).
Galperin, M. Y. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 (2006).
Parkinson, J. S. & Kofoid, E. C. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26, 71–112 (1992).
Stock, A. M., Robinson, V. L. & Goudreau, P. N. Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 (2000).
Wolanin, P. M., Thomason, P. A. & Stock, J. B. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3, REVIEWS3013 (2002).
Blacklow, S. C. & Kim, P. S. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat. Struct. Biol. 3, 758–762 (1996).
North, C. L. & Blacklow, S. C. Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry 39, 2564–2571 (2000).
Crimmins, D. L., Mische, S. M. & Denslow, N. D. Chemical cleavage of proteins in solution. Curr. Protoc. Protein Sci. Chapter 11, Unit 11.4 (2005).
Ni, J. & Kanai, M. Site-selective peptide/protein cleavage. Top Curr. Chem. 372, 103–123 (2016).
Hwang, P. M., Pan, J. S. & Sykes, B. D. Targeted expression, purification, and cleavage of fusion proteins from inclusion bodies in Escherichia coli. FEBS Lett. 588, 247–252 (2014).
Caldwell, T. A. et al. Low- q bicelles are mixed micelles. J. Phys. Chem. Lett. 9, 4469–4473 (2018).
Lata, S., Reichel, A., Brock, R., Tampe, R. & Piehler, J. High-affinity adaptors for switchable recognition of histidine-tagged proteins. J. Am. Chem. Soc. 127, 10205–10215 (2005).
Yang, J., Piai, A., Shen, H. B. & Chou, J. J. An exhaustive search algorithm to aid NMR-based structure determination of rotationally symmetric transmembrane oligomers. Sci. Rep. 7, 17373 (2017).
Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Mitra, K., Ubarretxena-Belandia, I., Taguchi, T., Warren, G. & Engelman, D. M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA 101, 4083–4088 (2004).
Sharpe, H. J., Stevens, T. J. & Munro, S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142, 158–169 (2010).
McMahon, H. T. & Boucrot, E. Membrane curvature at a glance. J. Cell. Sci. 128, 1065–1070 (2015).
Rossman, J. S., Jing, X., Leser, G. P. & Lamb, R. A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142, 902–913 (2010).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Guidotti, G. The composition of biological membranes. Arch. Intern. Med. 129, 194–201 (1972).
Trenker, R., Call, M. J. & Call, M. E. Progress and prospects for structural studies of transmembrane interactions in single-spanning receptors. Curr. Opin. Struct. Biol. 39, 115–123 (2016).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008).
Pielak, R. M., Oxenoid, K. & Chou, J. J. Structural investigation of rimantadine inhibition of the AM2-BM2 chimera channel of influenza viruses. Structure 19, 1655–1663 (2011).
Schnell, J. R. & Chou, J. J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451, 591–595 (2008).
OuYang, B. et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature 498, 521–525 (2013).
Wang, J., Pielak, R. M., McClintock, M. A. & Chou, J. J. Solution structure and functional analysis of the influenza B proton channel. Nat. Struct. Mol. Biol. 16, 1267–1271 (2009).
Szyperski, T., Neri, D., Leiting, B., Otting, G. & Wuthrich, K. Support of 1H NMR assignments in proteins by biosynthetically directed fractional 13C-labeling. J. Biomol. NMR 2, 323–334 (1992).
Shen, Y., Delaglio, F., Cornilescu, G. & Bax, A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009).
Acknowledgements
This work was supported by US National Institutes of Health grants GM116898 and AI127193 to J.J.C.
Author information
Authors and Affiliations
Contributions
Q.F., A.P., W.C., and J.J.C. conceived the study. K.X. performed the mass spectrometry analysis. Q.F., A.P., W.C., and J.J.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Fu, Q. et al. Mol. Cell 61, 602–613 (2016): https://doi.org/10.1016/j.molcel.2016.01.009
Piai, A., Devi, J., Fu, Q., & Chou, J. J. J. Am. Chem. Soc. 139, 18432–18435 (2017): https://doi.org/10.1021/jacs.7b09352
Chen, W. et al. Structure 26, 627–634.e4 (2018): https://doi.org/10.1016/j.str.2018.02.011
Fu, Q. et al. Proc. Natl. Acad. Sci. USA 115, E8892–E8899 (2018): https://doi.org/10.1073/pnas.1807259115
Supplementary information
Supplementary Software 1
ExSSO software for Linux.
Supplementary Software 2
ExSSO software for Mac OS.
Rights and permissions
About this article
Cite this article
Fu, Q., Piai, A., Chen, W. et al. Structure determination protocol for transmembrane domain oligomers. Nat Protoc 14, 2483–2520 (2019). https://doi.org/10.1038/s41596-019-0188-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0188-9
This article is cited by
-
Phosphatidylserine-dependent structure of synaptogyrin remodels the synaptic vesicle membrane
Nature Structural & Molecular Biology (2023)
-
Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation
Cell Research (2023)
-
Stabilization and structure determination of integral membrane proteins by termini restraining
Nature Protocols (2022)
-
HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes
Nature Chemical Biology (2020)
-
Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.