Abstract
Cancer cells subvert immune surveillance through inhibition of T cell effector function. Elucidation of the mechanism of T cell dysfunction is therefore central to cancer immunotherapy. Here, we report that dual specificity phosphatase 2 (DUSP2; also known as phosphatase of activated cells 1, PAC1) acts as an immune checkpoint in T cell antitumor immunity. PAC1 is selectively upregulated in exhausted tumor-infiltrating lymphocytes and is associated with poor prognosis of patients with cancer. PAC1hi effector T cells lose their proliferative and effector capacities and convert into exhausted T cells. Deletion of PAC1 enhances immune responses and reduces cancer susceptibility in mice. Through activation of EGR1, excessive reactive oxygen species in the tumor microenvironment induce expression of PAC1, which recruits the Mi-2β nucleosome-remodeling and histone-deacetylase complex, eventually leading to chromatin remodeling of effector T cells. Our study demonstrates that PAC1 is an epigenetic immune regulator and highlights the importance of targeting PAC1 in cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data and materials that support the findings of this study are available from the corresponding author upon reasonable request. The RNA-seq data in this study have been deposited in the SRA database with the accession code PRJNA512164. The ATAC-seq data have been deposited in the SRA database with the accession code PRJNA548971. The mass spectrometry proteome data in the FLAG pull-down assay have been deposited in the ProteomeXchange Consortium via the PRIDE (ref. 50) partner repository with the dataset identifier PXD012201 (username: reviewer61582@ebi.ac.uk, password: Dch8UNoq). The ChIP–seq data have been deposited in the GEO database with the accession code GSE141261. The CUT-Tag data have been deposited in the SRA database with the accession code PRJNA579783 Source data for Figs. 1–6 and Extended Data Figs. 1–6 are provided with the paper.
References
Elinav, E. et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 13, 759–771 (2013).
Lin, M. V., King, L. Y. & Chung, R. T. Hepatitis C virus-associated cancer. Annu. Rev. Pathol. 10, 345–370 (2015).
Maus, M. V. et al. Adoptive immunotherapy for cancer or viruses. Annu. Rev. Immunol. 32, 189–225 (2014).
Chen, L. & Flies, D. B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 (2013).
Levy, Y. et al. Enhanced T cell recovery in HIV-1-infected adults through IL-7 treatment. J. Clin. Invest. 119, 997–1007 (2009).
Ott, P. A., Hodi, F. S., Kaufman, H. L., Wigginton, J. M. & Wolchok, J. D. Combination immunotherapy: a road map. J. Immunother. Cancer. 5, 16 (2017).
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017).
Mahoney, K. M., Rennert, P. D. & Freeman, G. J. Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 14, 561–584 (2015).
Wykes, M. N. & Lewin, S. R. Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol. 18, 91–104 (2017).
Shen, E. et al. Chromatin remodeling by the NuRD complex regulates development of follicular helper and regulatory T cells. Proc. Natl Acad. Sci. USA 115, 6780–6785 (2018).
Lee, K. K. & Workman, J. L. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 (2007).
Torchy, M. P., Hamiche, A. & Klaholz, B. P. Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 72, 2491–2507 (2015).
Bornelov, S. et al. The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell 71, 56–72.e4 (2018).
Stecher, C. et al. PD-1 blockade promotes emerging checkpoint inhibitors in enhancing T cell responses to allogeneic dendritic cells. Front. Immunol. 8, 572 (2017).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Durgeau, A., Virk, Y., Corgnac, S. & Mami-Chouaib, F. Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Front. Immunol. 9, 14 (2018).
Gyorffy, B. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010).
Maglione, J. E. et al. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 61, 8298–8305 (2001).
Lang, P. A. et al. Reactive oxygen species delay control of lymphocytic choriomeningitis virus. Cell Death Differ. 20, 649–658 (2013).
van Stipdonk, M. J. et al. Dynamic programming of CD8+ T lymphocyte responses. Nat. Immunol. 4, 361–365 (2003).
Gett, A. V., Sallusto, F., Lanzavecchia, A. & Geginat, J. T cell fitness determined by signal strength. Nat. Immunol. 4, 355–360 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Rohan, P. J. et al. PAC-1: a mitogen-induced nuclear protein tyrosine phosphatase. Science 259, 1763–1766 (1993).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Vuzman, D. & Levy, Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 8, 47–57 (2012).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Shimbo, T. et al. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 9, e1004028 (2013).
Gorrini, C., Harris, I. S. & Mak, T. W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947 (2013).
Kalinowska, M., Bazdar, D. A., Lederman, M. M., Funderburg, N. & Sieg, S. F. Decreased IL-7 responsiveness is related to oxidative stress in HIV disease. PLoS One 8, e58764 (2013).
Wu, T. et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016).
Wang, Y. et al. Timing and magnitude of type I interferon responses by distinct sensors impact CD8 T cell exhaustion and chronic viral infection. Cell Host Microbe 11, 631–642 (2012).
Ghoneim, H. E. et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157.e19 (2017).
Dyson, H. J. Roles of intrinsic disorder in protein-nucleic acid interactions. Mol. Biosyst. 8, 97–104 (2012).
Lu, D. et al. The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates TH17 differentiation. Nat. Immunol. 16, 1263–1273 (2015).
Neufert, C., Becker, C. & Neurath, M. F. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2, 1998–2004 (2007).
McCausland, M. M. & Crotty, S. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods 147, 167–176 (2008).
Battegay, M. et al. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods 33, 191–198 (1991).
Gee, M. H. et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563.e16 (2018).
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21–29 (2015).
Wei, Z., Zhang, W., Fang, H., Li, Y. & Wang, X. esATAC: an easy-to-use systematic pipeline for ATAC-seq data analysis. Bioinformatics 34, 2664–2665 (2018).
Robinson, J. T. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Schmidt, D. et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 (2009).
Liu, X. et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Wang, G. et al. PTEN regulates RPA1 and protects DNA replication forks. Cell Res. 25, 1189–1204 (2015).
Huang, J. et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11, 592–603 (2009).
Vizcaino, J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 (2016).
Acknowledgements
We thank H. Tang (Institute Pasteur of Shanghai Chinese Academy of Sciences) for providing OT-I mice; J. Zhang (Peking University Health Science Center) for providing LCMV virus; Z. Zhu for help with mouse experiments; M. Chen, J. Gong and Z. Hou for help with tumor models; and X. Li. and Y. Zhu for help with bioinformatics analysis. This work was supported by grants from the National Key Research and Development Program of China (grant no. 2016YFA0500302 to Y.Y.), the National Natural Science Foundation of China (key grant nos 81430056, 31420103905 and 81874235 to Y.Y.; grant no. 81501360 to D.L.; and grant no. 31800749 to L.L.), the Beijing Natural Science Foundation (key grant no. 7161007 to Y.Y.), the Lam Chung Nin Foundation for Systems Biomedicine, the Fund for Fostering Young Scholars of Peking University Health Science Center (grant no. BMU2018YJ003 to D.L.) and the China Postdoctoral Science Foundation (grant no. 2018M630045 to L.L.).
Author information
Authors and Affiliations
Contributions
D.L., L.L. and Y.Y. conceived and designed the experiments. D.L. and L.L. performed most of the experiments and analyzed the data. Y.S., J.S., F.Q., Z.H. and Z.Y. assisted in experiments. Q.Y., Z.Z. and Y.J. provided technical assistance. Y.H., L.Z. and J.J. provided human samples. X.Z. did mass spectrometry analysis. G.Z. performed bioinformatics analysis. M.A.M. revised the manuscript. Y.Y. supervised this research. D.L., L.L., Y.S. and Y.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 PAC1 is correlated with inhibitory receptors in cancers.
(a) PAC1 (DUSP2) expression levels in various human tissues. Data are from the website (http://gemini.cancer-pku.cn/). (b) The relationship of disease-free survival of patients with hepatocellular carcinoma and inhibitory receptors (PDCD1, CTLA4, LAG3, HAVCR2, TIGIT, and CD160) expression. Data are from the TCGA database. (n = 325 samples, P = 0.0346). (c) The relationship of PAC1 expression with overall survival of lung cancer (n = 293 samples, P = 0.043), ovarian cancer (n = 55 samples, P = 0.03) or gastric cancer (n = 43 samples, P = 0.00044). Data are from the indicated GSE datasets. (d) The relationship of PAC1 expression with overall survival of colorectal carcinoma (n = 541 samples, P = 0.0047), esophageal carcinoma (n = 183 samples, P = 0.0046) or glioblastoma multiforme (n = 112 samples, P = 0.0175). Data are from the TCGA database. Statistical significance was assessed by Log-rank (Mantel-Cox) test (b-d).
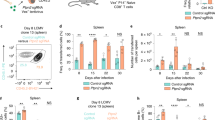
Extended Data Fig. 2 PAC1 mitigates T cell response.
(a) Flow cytometry analysis of cell cycle of Jurkat cells stably transfected with Mock or PAC1. (n = 3 cell cultures, mean ± s.e.m. * P = 0.0226, ** P = 0.0015 and **** P = 0.0000012). (b) qRT-PCR analysis of mRNA levels of GZMB, IFNG, TNF and IL2 in Jurkat cells stably expressing Mock, PAC1WT or PAC1C/S left untreated (UT) or stimulated with PMA plus ionomycin (PMA + iono) for 6 h (n = 2 cell cultures). (c) Flow cytometry analysis of the frequency of CD8+ and CD4+ T cells from wild-type or Pac1-/- LN stimulated with anti-CD3 (2 μg/ml) plus anti-CD28 (1 μg/ml) for the indicated lengths of time. The CD8+/CD4+ T cell ratio was calculated (n = 4 mice, mean ± s.e.m. ** P = 0.0075 and **** P < 0.0001). (d) Wild-type and Pac1-/- CD4+ or CD8+ T cells were treated with anti-CD3 plus anti-CD28 for 48 h, and proliferation was determined by CFSE dilution assay (n = 4 mice, mean ± s.e.m. NS = 0.1172, * P < 0.05, ** P < 0.01 and **** P = 0.000093). (e) Naive CD8+ T cells from wild-type or Pac1-/- mice were treated with anti-CD3 plus anti-CD28 for 24 h, followed by quantification of IL-2 production by ELISA assay (pooled mice, n = 6 cell cultures, mean ± s.e.m. **** P = 0.0000003) or Il2 expression by qRT-PCR analysis (pooled mice, n = 2 cell cultures, mean ± s.e.m. NS = 0.6353 and *** P = 0.0004). Statistical significance was assessed by two-tailed unpaired Student’s t test (a,c,d,e). Data are representative of three (b-d) or two (a,e) independent experiments.
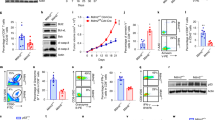
Extended Data Fig. 3 PAC1 impedes host anti-tumor immunity.
(a) Body weight changes of mice followed with AOM-DSS treatment (n = 5 mice, mean ± s.e.m. * P = 0.015, ** P < 0.01 and *** P < 0.001). (b) Hematoxylin-and-eosin (H&E) staining of tumors in indicated mice (n = 7 mice). Scale bars, 50 μm. (c) Tumor growth of mice subcutaneously injected with 1 × 106 B16-F10 cells (WT, n = 7 mice; Pac1-/-, n = 6 mice, mean ± s.e.m. * P < 0.05, ** P = 0.0092 and *** P = 0.00062). (d) Survival comparison of indicated mice (c) (WT, n = 7 mice; Pac1-/-, n = 6 mice, ** P = 0.0022). (e) qRT-PCR analysis of Pac1 mRNA levels in naive CD8+ T cells left untreated or activated with anti-CD3 plus anti-CD28 for 2 or 48 hours, T cells purified from MLN and melanoma and the B16 cell line (pooled mice, n = 2 technical replicates, mean ± s.e.m. *** P < 0.001 and **** P = 0.000014). (f, g) Indicated mononuclear cells stimulated with PMA plus ionomycin for 5 h (f) or left untreated (g). IFN-γ and TNF (f) or PD-1 (g) expression in CD8+ cells was evaluated with flow cytometry (WT, n = 5 mice; Pac1-/-, n = 6 mice, mean ± s.e.m. NS = 0.2568, * P = 0.018, ** P < 0.01, *** P = 0.00014 and **** P = 0.0000077). (h) Gross appearance of lungs and quantification of lung metastases from indicated mice 20 days after intravenous injection with 1 × 106 B16-F10 cells (n = 5 mice, mean ± s.e.m. * P = 0.0398). Statistical significance was assessed by the Log-rank (Mantel-Cox) test (d) or two-tailed unpaired Student’s t test (a,c,e-h). Data are representative of three (c,d,h) or two (a,b,e-g) independent experiments.
Extended Data Fig. 4 PAC1 is essential for ROS-mediated T cell dysfunction.
(a) Morphological examination of lungs from indicated mice on day 7 post-infection with LCMV Cl13 (n = 4 mice). (b) H&E staining of the lung from indicated mice untreated (UT) or stimulated LCMV Cl13 for 7 days (n = 4 mice). Scale bars, 100 μm. (c) The frequencies of CD8+ GP33-41+ cells isolated from the lung of indicated mice on day 7 post-infection with LCMV Cl13 (n = 5 mice, mean ± s.e.m. ** P = 0.0059). (d) Flow cytometry analysis of the expression of IFN-γ and TNF in CD8+ GP33-41+ cells stimulated with GP33-41 (2 μg/ml) peptides (n = 5 mice, mean ± s.e.m. *** P < 0.001). (e) The splenocytes from indicated mice immunized by LCMV Arm were incubated with LLC cells infected with LCMV Arm (left) or Cl13 (right). The cell viability was measured by CCK8 (n = 3 cell cultures, mean ± s.e.m. * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P = 0.000044). (f) GSEA of genes expressed in indicated CD8+ T cells on day 7 post-infection with LCMV Cl13 (n = 2 mice). KO, Pac1-/-; ES, enrichment score; NES, normalized enrichment score. (g, h) Wild-type and Pac1-/- lymphocytes were treated with anti-CD3 (2 μg/ml) and anti-CD28 (1 μg/ml) in the presence of H2O2 (50 μM) for 6 (g) or 24 h (h), followed by flow cytometric analysis of TNF expression (g) or CD25 expression (h) in CD8+ T cells respectively (n = 3 mice, mean ± s.e.m. NS = 0.1609, ** P = 0.0024, *** P = 0.0002 and **** P = 0.000067). Statistical significance was assessed by two-tailed unpaired Student’s t test (c,d,e-h). Data are representative of three (a-d) or two (e-h) independent experiments.
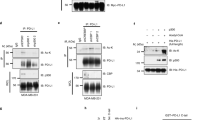
Extended Data Fig. 5 PAC1 interacts with NuRD complex and modulates chromatin accessibility.
(a) DAVID enrichment analysis for PAC1 interactome from Jurkat cells treated with PMA plus ionomycin for 4 h. (b) Co-immunoprecipitation analysis of PAC1-GFP with indicated components of NuRD complex. (c) Co-immunoprecipitation analysis of PAC1WT-GFP or PAC1C/S-GFP together with FLAG-tagged HDAC1 (left panel) or HDAC2 (right panel). (d) Co-immunoprecipitation analysis of HDAC1-HA together with FLAG-tagged PAC1 and its truncations. (e) Flow cytometry analysis of GZMB expression in Jurkat cells stably expressing indicated vectors with PMA plus ionomycin in the presence or absence of HDAC inhibitor Trichostatin A (TSA) or Sodium Butyrate (SB) for 6 h (n = 6 cell cultures, mean ± s.e.m.). (f) qRT-PCR analysis of mRNA levels of TNF, IL2, GZMB or IFNG in Jurkat cells stably expressing Mock, PAC1WT or PAC1C/S left untreated or stimulated with PMA plus ionomycin in the presence or absence of TSA or SB for 6 h (n = 2 cell cultures). (g) Secondary spectra of acetylated lysine 5 and 8 residues of Histone H4 (top panel) as well as acetylated lysine 18 and 23 residues of Histone H3 (bottom panel) affected by PAC1WT or PAC1C/S during T cell activation. (h) Immunoblot analysis of acetyl-H3K27 in Jurkat cells stably expressing indicated vectors left treated with PMA plus ionomycin or not. (i-k) Distribution of chromatin regions or peaks in genomic regions of the ATAC-seq (i), ChIP-seq (j) and CUT&Tag (k) as in Fig. 6f. Statistical significance was assessed by two-tailed unpaired Student’s t test (a,e). Data are representative of two independent experiments (a-k).
Extended Data Fig. 6 Model for the role of PAC1 in ROS-mediated tumor-infiltrating lymphocyte dysfunction.
Prolonged antigenic stimulation induces excessive ROS production, which in turn upregulates PAC1 expression via EGR1 signaling in effector T cells. Instead of utilizing phosphatase activity, PAC1 dampens T cell cytotoxic function via recruitment of the NuRD complex and remodeling the epigenetic program of effector T cells, eventually leading to T cell exhaustion and cancer immune escape.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1a–c,e,f.
Source Data Fig. 1
Unprocessed western blots for Fig. 1d.
Source Data Fig. 2
Statistical source data for Fig. 2b,d,f,h,i.
Source Data Fig. 3
Statistical source data for Fig. 3a–c,e,g–i.
Source Data Fig. 3
Unprocessed western blots for Fig. 3d.
Source Data Fig. 4
Statistical source data for Fig. 4a–f.
Source Data Fig. 4
Unprocessed western blots for Fig. 4b.
Source Data Fig. 5
Statistical source data for Fig. 5b,f.
Source Data Fig. 5
Unprocessed western blots for Fig. 5b,d.
Source Data Fig. 6
Statistical source data for Fig. 6d,e.
Source Data Fig. 6
Unprocessed western blots for Fig. 6b,d.
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1b,d.
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2a–e.
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3a,c–h.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4c–e,g,h.
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5a,e,f.
Source Data Extended Data Fig. 5
Unprocessed western blots for Extended Data Fig. 5b–d,h.
Rights and permissions
About this article
Cite this article
Dan Lu, Liu, L., Sun, Y. et al. The phosphatase PAC1 acts as a T cell suppressor and attenuates host antitumor immunity. Nat Immunol 21, 287–297 (2020). https://doi.org/10.1038/s41590-019-0577-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0577-9
This article is cited by
-
Id2 epigenetically controls CD8+ T-cell exhaustion by disrupting the assembly of the Tcf3-LSD1 complex
Cellular & Molecular Immunology (2024)
-
Single-cell transcriptome analysis profiling lymphatic invasion-related TME in colorectal cancer
Scientific Reports (2024)
-
The transcription factor AP2XI-2 is a key negative regulator of Toxoplasma gondii merogony
Nature Communications (2024)
-
Challenges and new technologies in adoptive cell therapy
Journal of Hematology & Oncology (2023)
-
Critical role of VHL/BICD2/STAT1 axis in crystal-associated kidney disease
Cell Death & Disease (2023)