Abstract
Targeting the p53–MDM2 pathway to reactivate tumor p53 is a chemotherapeutic approach. However, the involvement of this pathway in CD8+ T cell-mediated antitumor immunity is unknown. Here, we report that mice with MDM2 deficiency in T cells exhibit accelerated tumor progression and a decrease in tumor-infiltrating CD8+ T cell survival and function. Mechanistically, MDM2 competes with c-Cbl for STAT5 binding, reduces c-Cbl-mediated STAT5 degradation and enhances STAT5 stability in tumor-infiltrating CD8+ T cells. Targeting the p53–MDM2 interaction with a pharmacological agent, APG-115, augmented MDM2 in T cells, thereby stabilizing STAT5, boosting T cell immunity and synergizing with cancer immunotherapy. Unexpectedly, these effects of APG-115 were dependent on p53 and MDM2 in T cells. Clinically, MDM2 abundance correlated with T cell function and interferon-γ signature in patients with cancer. Thus, the p53–MDM2 pathway controls T cell immunity, and targeting this pathway may treat patients with cancer regardless of tumor p53 status.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Single-cell RNA-seq data that support the findings of this study were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus under the accession number GSE72056. All other data that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Wade, M., Li, Y. C. & Wahl, G. M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13, 83–96 (2013).
Chene, P. Inhibiting the p53–MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer 3, 102–109 (2003).
Shangary, S. & Wang, S. Small-molecule inhibitors of the MDM2–p53 protein–protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu. Rev. Pharm. Toxicol. 49, 223–241 (2009).
Khoo, K. H., Verma, C. S. & Lane, D. P. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 (2014).
Karni-Schmidt, O., Lokshin, M. & Prives, C. The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617–644 (2016).
Zhang, Z., Li, M., Wang, H., Agrawal, S. & Zhang, R. Antisense therapy targeting MDM2 oncogene in prostate cancer: effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc. Natl Acad. Sci. USA 100, 11636–11641 (2003).
Topalian, S. L., Drake, C. G. & Pardoll, D. M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015).
Zou, W., Wolchok, J. D. & Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv324 (2016).
Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018).
Gajewski, T. F., Schreiber, H. & Fu, Y. X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 (2013).
Zou, W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer 5, 263–274 (2005).
Kawashima, H. et al. Tumor suppressor p53 inhibits systemic autoimmune diseases by inducing regulatory T cells. J. Immunol. 191, 3614–3623 (2013).
Pan, Y. & Haines, D. S. The pathway regulating MDM2 protein degradation can be altered in human leukemic cells. Cancer Res. 59, 2064–2067 (1999).
Komarov, P. G. et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285, 1733–1737 (1999).
Waldmann, T. A. & Chen, J. Disorders of the JAK/STAT pathway in T cell lymphoma pathogenesis: implications for immunotherapy. Annu. Rev. Immunol. 35, 533–550 (2017).
O'Shea, J. J., Gadina, M. & Schreiber, R. D. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109, S121–S131 (2002).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Goh, E. L., Zhu, T., Leong, W. Y. & Lobie, P. E. c-Cbl is a negative regulator of GH-stimulated STAT5-mediated transcription. Endocrinology 143, 3590–3603 (2002).
Dieudonne, F. X. et al. Promotion of osteoblast differentiation in mesenchymal cells through Cbl-mediated control of STAT5 activity. Stem Cells 31, 1340–1349 (2013).
Aguilar, A. et al. Discovery of 4-((3′R,4′S,5′R)-6″-chloro-4′-(3-chloro-2-fluorophenyl)-1′-ethyl-2″-oxodispiro[cyclohexane-1,2′-pyrrolidine-3′,3″-indoline]-5′-carboxamido)bicyclo[2.2.2]octane-1-carboxylic acid (AA-115/APG-115): a potent and orally active murine double minute 2 (MDM2) inhibitor in clinical development. J. Med. Chem. 60, 2819–2839 (2017).
Brown, C. J., Lain, S., Verma, C. S., Fersht, A. R. & Lane, D. P. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer 9, 862–873 (2009).
Watanabe, M., Moon, K. D., Vacchio, M. S., Hathcock, K. S. & Hodes, R. J. Downmodulation of tumor suppressor p53 by T cell receptor signaling is critical for antigen-specific CD4+ T cell responses. Immunity 40, 681–691 (2014).
Banerjee, A. et al. Lack of p53 augments antitumor functions in cytolytic T cells. Cancer Res. 76, 5229–5240 (2016).
Lujambio, A. et al. Non-cell-autonomous tumor suppression by p53. Cell 153, 449–460 (2013).
Tsai, N. P. et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 151, 1581–1594 (2012).
Fang, D. D. et al. MDM2 inhibitor APG-115 synergizes with PD-1 blockade through enhancing antitumor immunity in the tumor microenvironment. J. Immunother. Cancer 7, 327 (2019).
Itahana, K. et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12, 355–366 (2007).
Li, Y. et al. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem. 62, 448–466 (2019).
Lin, H. et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J. Clin. Invest. 128, 805–815 (2018).
Wang, W. et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274 (2019).
Grier, J. D., Yan, W. & Lozano, G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis 32, 145–147 (2002).
Wang, D. et al. Controllable synthesis of dual-MOFs nanostructures for pH-responsive artemisinin delivery, magnetic resonance and optical dual-model imaging-guided chemo/photothermal combinational cancer therapy. Biomaterials 100, 27–40 (2016).
Huang, M. et al. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods 15, 539–542 (2018).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We thank Y. Sun for providing the His–Ub and His–Ub K48R plasmids, W. Wei for providing the HA–MDM2 plasmid, F. Hua for providing the HA–c-Cbl plasmid and G. Lozano for providing the Mdm2flox/flox mice. We are grateful to Y. Sun and W. Wei for scientific input. This work was supported in part by research grants from the NIH/NCI (CA248430, CA217648, CA123088, CA099985, CA193136 and CA152470 to W.Z.) and from the NIH through University of Michigan Rogel Cancer Center support grant P30CA46592.
Author information
Authors and Affiliations
Contributions
J.Z. and W.Z. conceived the project and designed the experiments. J.Z. performed most of the experiments with help from all authors. S. Wei and S.G. generated the Mdm2+/+Cd4-Cre, Mdm2fl/flCd4-Cre, p53+/+Cd4-Cre and p53fl/flCd4-Cre mice. J.Y., Y.Y., H.L., G.L., W.D., Weichao Wang, X. Lang and L.V. assisted with the tumor experiments. I.K. and L.V. assisted with the flow cytometry analysis. S.L., X. Li and J.L. performed the bioinformatics analysis. A.A. and S. Wang provided APG-115. S. Wang, P.L., Weimin Wang and I.K. provided scientific input and discussions. J.Z., I.K. and W.Z. wrote the manuscript. W.Z. supervised the work and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The University of Michigan has filed patent applications on APG-115 and its analogs, which have been licensed to Ascentage Pharma Group International for clinical development. S. Wang is a co-founder and paid consultant of Ascentage Pharma Group International and owns stock in Ascentage. The University of Michigan also owns stock in Ascentage.
Additional information
Peer review information Nature Immunology thanks Ping-Chih Ho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
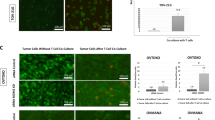
Extended Data Fig. 1 MDM2 controls CD8+ T cell-mediated anti-tumor immunity.
a, b, Effect of TCR-engagement on MDM2 in CD8+ T cells. Human (a) and mouse (b) CD8+ T cells were stimulated with anti-CD3 and anti-CD28 mAbs. Expression of MDM2 was determined by Western blotting. mean ± SEM, n = 5, *p < 0.05, **p < 0.01, ****p < 0.0001, one-way ANOVA. c, d, Protein levels of MDM2 in T cells from Mdm2+/+Cd4-Cre and Mdm2fl/flCd4-Cre mice. Results are expressed as the relative mean density of MDM2 ± SEM, n = 6, **p < 0.01, Mann Whitney test. e–j, Phenotype of Mdm2fl/flCd4-Cre mice. The representative images show the body size (e) and the lymphoid organs (f) of Mdm2+/+Cd4-Cre and Mdm2fl/flCd4-Cre mice. The percentages of CD8+ T cells in T cells (g), the representative images of flow dot plots (h), PD-1+ and CD25+ cells in T cell subsets (i) in different organs, and T cells in CD45+ immune cells (j) were analyzed by FACS. k, l, Effect of MDM2 deficiency on T cell proliferation under the homeostatic stimulation. Mdm2+/+ and Mdm2–/– CD8+ T cells were cultured with IL-7 and IL-15 in response to PMA. Ki67 expression was analyzed by FACS. mean ± SEM, n = 5, **p < 0.05, Mann Whitney test. m–q, Role of MDM2 deficiency in ID8 tumor infiltrating CD8+ T cells. The anti-apoptotic (Bcl2 and Bcl-xL) and pro-apoptotic (Bak, cl-caspase 8, and cl-caspase 3) proteins were detected in tumor infiltrating CD8+ T cells by Western blotting. Results are expressed as the relative mean density of specific proteins ± SEM, n = 4, *p < 0.05, Mann Whitney test. r–t, Effect of MDM2 deficiency on MC38 tumor progression and T cell proliferation. MC38 tumors were inoculated into Mdm2+/+Cd4-Cre and Mdm2fl/flCd4-Cre mice. Tumor weight was measured on day 21 (r). The percentages of Ki67+ cells in tumor infiltrating CD8+ T cells were analyzed by FACS (s-t). mean ± SEM, n = 4 – 5, *p < 0.05, **p < 0.01, Mann Whitney test. u. Protein levels of p53 in T cells isolated from p53+/+Cd4-Cre and p53fl/flCd4-Cre mice. One of 6 experiments is shown. v–w, Phenotype of p53fl/flCd4-Cre mice. The representative images show the body size (v) and lymphoid organs (w) of p53+/+Cd4-Cre and p53fl/flCd4-Cre mice. x, T cell subset distribution in p53+/+Cd4-Cre and p53fl/flCd4-Cre mice. One of 3 representative dot-plots is shown.
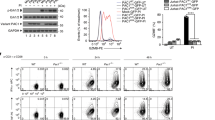
Extended Data Fig. 2 MDM2 regulates STAT5 protein expression in T cells.
a–h, Effect of MDM2 knockdown on MDM2, STAT, and JAK expression. LS 174T cells were transfected with 3 different shRNAs against Mdm2 or control. The relevant proteins were detected by immunoblots. Results are expressed as the relative mean density of specific protein ± SEM, n = 3, ****p < 0.0001, one-way ANOVA. i-j, Effect of Mdm2 deficiency on Stat5 transcripts. Real time PCR showed Stat5α and Stat5β mRNAs in Mdm2+/+ and Mdm2–/– T cells. Five biological replicates.
Extended Data Fig. 3 MDM2 prevents c-Cbl-mediated STAT5 degradation.
a, b, Immunoblots showed STAT5 in Jurkat T cells in the presence of MG132. mean ± SEM, n = 5, **p < 0.01, Mann Whitney test. c, Effect of MDM2 on STAT5 protein expression. Wild-type and shMdm2-LS 174T cells were treated with or without MG132. STAT5 levels were measured by Western blotting. Results are expressed as the relative mean density of STAT5 ± SEM, n = 3, *p < 0.05, ***p < 0.001, one way-ANOVA. d–i, Effect of c-Cbl on STAT5 and MDM2 expression. LS 174T and Jurkat T cells were transfected with 2 shRNAs against c-Cbl or control. c-Cbl (d-e), STAT5 (f-g), and MDM2 (h-i) expression was detected by Western blotting. Results are expressed as the relative mean density of specific protein ± SEM, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA. j, k, Effect of c-Cbl and MDM2 on T cell proliferation. Activated mouse Mdm2–/– CD8+ T cells (j) and shMdm2-Jurkat T (k) cells were transfected with si-c-Cbl. Ki67 expression was determined by FACS. mean ± SEM, n = 3, **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA. l–n, Effect of MDM2 on c-Cbl expression in Jurkat T and LS 174T cells. Jurkat T and LS 174T cells were transfected with shRNAs against Mdm2 or control. Immunoblots showed c-Cbl expression. Results are expressed as the relative mean density of c-Cbl ± SEM, n = 3, NS, not significant. o-p, Role of MDM2 deficiency in mouse T cells. CD3+ T cells were isolated from Mdm2+/+Cd4-Cre and Mdm2fl/flCd4-Cre mice. STAT5 (o) and c-Cbl (p) expression were determined by Western blotting. Results are expressed as the relative mean density of specific protein ± SEM, n = 4, *p < 0.05, Mann Whitney test. q, Role of MDM2 in STAT5 expression. c-Cbl+/+ and c-Cbl–/– LS 174T cells were transfected with HA-MDM2 and empty vector. STAT5 levels were determined by Western blotting. Results are expressed as the relative mean density of STAT5 ± SEM, n = 3, *p < 0.05, **p < 0.01, one-way ANOVA.
Extended Data Fig. 4 Targeting p53-MDM2 interaction elicits tumor p53 independent anti-tumor activity.
a, APG115 structure is shown. b–e, Effect of APG115 on MDM2 and p53 expression in T cells. Human (b-c) and mouse (d-e) T cells were stimulated with anti-CD3 and anti-CD28 mAbs in the presence of different concentrations of APG115. MDM2 (b, d) and p53 (c, e) expression was determined by Western blotting. Results are expressed as the relative mean density of MDM2 and p53 ± SEM, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA. f–h, Effect of APG115 on p53 wild-type tumor cells. B16F10 (f), CT26 (g) and ID8 (h) cells were treated with different concentrations of APG115. Immunoblots showed MDM2, p53, and p21 expression. i, Effect of APG115 on p53 mutant tumor cells. MC38 cells were treated with different concentrations of APG115. Immunoblots showed PARP1, p53, and MDM2 expression. j, Effect of APG115 on p53-null 4T1 tumor cells. 4T1 tumor cells were treated with different concentrations of APG115. Immunoblots showed PARP1, p53, and MDM2 expression. k, Effect of APG115 on p53–/–CD8+ T cell survival. p53–/–CD8+ T cells were treated with APG115. Annexin V+ expression was analyzed by FACS. mean ± SEM, n = 4, NS, not significant.
Extended Data Fig. 5 Targeting p53-MDM2 interaction activates CD8+ T cell-mediated anti-tumor immunity.
a, b, Effect of APG115 on tumor growth in NSG mice. MC38 tumor bearing NSG mice were treated with APG115. Tumor weight (a) and image (b) are shown after 21 days. mean ± SEM, Vehicle, n = 9; APG115, n = 11, NS, not significant. c, d, Effect of CD4+ T cell depletion on APG15-mediated anti-tumor activity. MC38 tumor bearing C57BL/6J mice were treated with APG115 and/or anti-CD4. (c) Tumor volume was monitored. mean ± SEM, n = 6. **** p < 0.0001, two-way ANOVA. (d) The efficacy of CD4+ T cell depletion. Tumor draining lymph node (TDLN) T cells were analyzed by FACS. One of 5 dot-plots is shown. e, f, Role of APG115 on 4T1 tumor infiltrating CD8+ T cells. The percentages of IL-2+ and tumor necrosis factor-α (TNFα)+ cells in CD8+ T cells were determined by FACS. mean ± SEM, n = 5, *p < 0.05, **p < 0.01, Mann Whitney test. g, h, Effect of MDM2 on APG115-regulated T cell apoptosis. B16F10 tumor bearing Mdm2+/+Cd4-Cre or Mdm2fl/flCd4-Cre mice were treated with APG115. The percentages of Annexin V+ cells in tumor infiltrating CD8+ T cells were determined by FACS. mean ± SEM, n = 4–6; *p < 0.05, one-way ANOVA.
Extended Data Fig. 6 Targeting p53-MDM2 interaction stabilizes T cell STAT5 and synergizes with immunotherapy.
a–e, Role of APG115 in CD8+ T cells. (a, c) Mouse Mdm2+/+Cd4-Cre and Mdm2fl/flCd4-Cre CD8+ T cells were stimulated with anti-CD3 and anti-CD28 in the presence of APG115. Western blots showed STAT5 (a), Bcl-xL (b), and cl-caspase 8 (c) in T cells. mean ± SEM, n = 3, *p < 0.05, **p < 0.01, ****p < 0.0001, one-way ANOVA, (d) Western blots showed expression of STATs and MDM2 in MC38 tumor infiltrating CD8+ T cells in mice treated with APG115. mean ± SEM, n = 6, ***p < 0.001, one-way ANOVA. (e) Western blots showed STAT5 expression in human CD8+ T cells stimulated with anti-CD3 and anti-CD28 in the presence of APG115. mean ± SEM, n = 4, *p < 0.05, Mann Whitney test. f–h, Role of APG115 in CD8+ T cells. Mouse CD8+ T cells were stimulated with anti-CD3 and anti-CD28 in the presence of APG115. IFNγ+ (f), TNFα+ (g), and IL-2+ (h) cells in CD8+ T cells were analyzed by FACS. mean ± SEM, n = 4, *p < 0.05, Mann Whitney test. i. Effect of STAT5CA expression on MDM2-mediated T cell survival. Activated Mdm2+/+ and Mdm2–/– CD8+ cells were transduced with STAT5CA or vector expressing retrovirus, and treated with APG115. The percentages of Annexin V+ cells in Mdm2+/+ or Mdm2–/– CD8+ T cells were determined by FACS. mean ± SEM, n = 4, *p < 0.05, ****p < 0.0001, one-way ANOVA. j. Effect of STAT5 knock down in OT-I-mediated tumor killing. B16F10-OVA cells were cultured with STAT5 deficiency OT-I T cells for 24 hours. Tumor cell apoptosis was determined by flow cytometry analysis. Results are shown as the percentages of 7AAD+ tumor cells. mean ± SEM, n = 4, ***p < 0.001, one-way ANOVA. k, Role of APG115 on MDM2, c-Cbl, and STAT5 expression in p53–/– T cells. p53–/–CD8+ T cells were treated with APG115. MDM2, c-Cbl, and STAT5 levels were analyzed by Western Blotting. One of 3 experiments is shown.
Extended Data Fig. 7 Mdm2 expression correlates with immune gene signatures in patients with cancer.
a–c, GSEA analysis showed differentially expressed gene signatures - including T cell receptor signaling pathway (a), T cell activation (b), and cytokine production regulation (c) - in human melanoma infiltrating T cells with high vs low Mdm2 expression. d, Schematic diagram illustrates that MDM2 competes with c-Cbl and prevents c-Cbl binding to STAT5, thereby reducing c-Cbl-mediated STAT5 degradation and supporting CD8+ T cell-mediated immunity and immunotherapy.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots,
Source Data Extended Data Fig. 5
Statistical source data,
Source Data Extended Data Fig. 6
Statistical source data,
Source Data Extended Data Fig. 6
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Zhou, J., Kryczek, I., Li, S. et al. The ubiquitin ligase MDM2 sustains STAT5 stability to control T cell-mediated antitumor immunity. Nat Immunol 22, 460–470 (2021). https://doi.org/10.1038/s41590-021-00888-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-00888-3
This article is cited by
-
Targeting MDM2 in malignancies is a promising strategy for overcoming resistance to anticancer immunotherapy
Journal of Biomedical Science (2024)
-
JAK/STAT in leukemia: a clinical update
Molecular Cancer (2024)
-
Genome-wide CRISPR screen identifies ESPL1 limits the response of gastric cancer cells to apatinib
Cancer Cell International (2024)
-
Pharmacological reactivation of p53 in the era of precision anticancer medicine
Nature Reviews Clinical Oncology (2024)
-
Translating p53-based therapies for cancer into the clinic
Nature Reviews Cancer (2024)