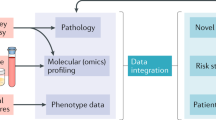
Abstract
A huge array of data in nephrology is collected through patient registries, large epidemiological studies, electronic health records, administrative claims, clinical trial repositories, mobile health devices and molecular databases. Application of these big data, particularly using machine-learning algorithms, provides a unique opportunity to obtain novel insights into kidney diseases, facilitate personalized medicine and improve patient care. Efforts to make large volumes of data freely accessible to the scientific community, increased awareness of the importance of data sharing and the availability of advanced computing algorithms will facilitate the use of big data in nephrology. However, challenges exist in accessing, harmonizing and integrating datasets in different formats from disparate sources, improving data quality and ensuring that data are secure and the rights and privacy of patients and research participants are protected. In addition, the optimism for data-driven breakthroughs in medicine is tempered by scepticism about the accuracy of calibration and prediction from in silico techniques. Machine-learning algorithms designed to study kidney health and diseases must be able to handle the nuances of this specialty, must adapt as medical practice continually evolves, and must have global and prospective applicability for external and future datasets.
Key points
-
Big data in nephrology can provide essential information about kidney disease burden, molecular mechanisms, novel risk factors and therapeutic targets.
-
Artificial intelligence and machine-learning approaches that utilize big data could be used for a variety of applications in nephrology, including early diagnosis and prognosis, as well as clinical decision-support systems for personalized selection of therapy.
-
Data curation and standardization enable interoperability, facilitate consolidation and exchange of high-quality data from different sources, create independence from manufacturers and ease competition as comparable products are offered by all market players.
-
Sources of big data in nephrology include patient registries, population surveys, electronic health records, open-access clinical trials, mobile health devices and molecular data repositories.
-
Large-scale acquisition of annotated molecular and clinical data, together with advances in machine learning approaches, open-source computational packages, affordable computation power and cloud storage, will all facilitate more novel data-driven approaches in nephrology.
-
Challenges for the utilization of big data in nephrology include issues relating to data governance and protection, siloed datasets, data heterogeneity, small sample sizes and a lack of consistent research funding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Erickson, K. F., Qureshi, S. & Winkelmayer, W. C. The role of big data in the development and evaluation of US dialysis care. Am. J. Kidney Dis. 72, 560–568 (2018).
Adimadhyam, S. et al. Leveraging the capabilities of the FDA’s sentinel system to improve kidney care. J. Am. Soc. Nephrol. 31, 2506–2516 (2020).
Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016).
Escobar, G. J. et al. Automated identification of adults at risk for in-hospital clinical deterioration. N. Engl. J. Med. 383, 1951–1960 (2020).
Hulsen, T. et al. From big data to precision medicine. Front. Med. 6, 34 (2019).
Cahan, E. M., Hernandez-Boussard, T., Thadaney-Israni, S. & Rubin, D. L. Putting the data before the algorithm in big data addressing personalized healthcare. NPJ Digit. Med. 2, 78 (2019).
Liu, F. X., Rutherford, P., Smoyer-Tomic, K., Prichard, S. & Laplante, S. A global overview of renal registries: A systematic review Epidemiology and Health Outcomes. BMC Nephrol. 16, 1–10 (2015).
Bikbov, B. et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 395, 709–733 (2020).
Friedman, D. J., Parrish, R. G. & Ross, D. A. Electronic health records and US public health: Current realities and future promise. Am. J. Public Health 103, 1560–1567 (2013).
Murphy, D. et al. Trends in prevalence of chronic kidney disease in the United States. Ann. Intern. Med. 165, 473–481 (2016).
Chan, L. et al. The effect of depression in chronic hemodialysis patients on inpatient hospitalization outcomes. Blood Purif. 43, 226–234 (2017).
Cheung, A. et al. Impact of atrial fibrillation in patients with chronic kidney disease undergoing transcatheter aortic valve replacement: Insights of the Healthcare Cost and Utilization Project’s National Inpatient Sample. Cardiovasc. Revasc. Med. 19, 21–25 (2018).
Matsushita, K. et al. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet 375, 2073–2081 (2010).
Zoccali, C., Brancaccio, D. & Nathan, M. J. Causality at the dawn of the ‘omics’ era in medicine and in nephrology. Nephrol. Dial. Transplant. 31, 1381–1385 (2016).
Weber, G. M., Mandl, K. D. & Kohane, I. S. Finding the missing link for big biomedical data. J. Am. Med. Assoc. 311, 2479–2480 (2014).
Nadkarni, G. N., Coca, S. G. & Wyatt, C. M. Big data in nephrology: promises and pitfalls. Kidney Int. 90, 240–241 (2016).
Pezoulas, V. C. et al. Medical data quality assessment: on the development of an automated framework for medical data curation. Comput. Biol. Med. 107, 270–283 (2019).
Danese, M. D., Halperin, M., Duryea, J. & Duryea, R. The generalized data model for clinical research. BMC Med. Inform. Decis. Mak. 19, 1–13 (2019).
Fleurence, R. L. et al. Launching PCORnet, a national patient-centered clinical research network. J. Am. Med. Inform. Assoc. 21, 578–582 (2014).
Murphy, S. N. et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J. Am. Med. Inform. Assoc. 17, 124–130 (2010).
Klann, J. G., Joss, M. A. H., Embree, K. & Murphy, S. N. Data model harmonization for the all of us research program: transforming i2b2 data into the OMOP common data model. PLoS One 14, 1–13 (2019).
Kush, R. D. et al. FAIR data sharing: the roles of common data elements and harmonization. J. Biomed. Inform. 107, 103421 (2020).
Wilkinson, M. D. et al. Comment: the FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 1–9 (2016).
Kubben, P., Dumontier, M. & Dekker, A. Fundamentals of clinical data science. (Springer, 2019).
Dreyer, N. A. & Garner, S. Registries for robust evidence. JAMA 302, 790–791 (2009).
Jager, K. J. & Wanner, C. Fifty years of ERA-EDTA registry — a registry in transition. Kidney Int. Suppl. 5, 12–14 (2015).
Choi, N. G., Sullivan, J. E., DiNitto, D. M. & Kunik, M. E. Health care utilization among adults with CKD and psychological distress. Kidney Med. 1, 162–170 (2019).
Robinson, B. M., Bieber, B., Pisoni, R. L. & Port, F. K. Dialysis outcomes and practice patterns study (DOPPS): Its strengths, limitations, and role in informing practices and policies. Clin. J. Am. Soc. Nephrol. 7, 1897–1905 (2012).
DOPPS. DPM sampling, study design, and calculation methods. DOPPS https://www.dopps.org/DPM/Data_Sources_Methods.pdf (2020).
Dienemann, T. et al. International Network of Chronic Kidney Disease cohort studies (iNET-CKD): a global network of chronic kidney disease cohorts. BMC Nephrol. 17, 1–9 (2016).
Saran, R. et al. US Renal Data System 2019 Annual Data Report: epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 75, A6–A7 (2020).
Go, A. S., Chertow, G. M., Fan, D., McCulloch, C. E. & Hsu, C. Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med. 351, 1296–1305 (2004).
Saran, R. et al. US Renal Data System 2014 Annual Data Report: epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 66, A7 (2015).
Mendu, M. L. et al. Development of an electronic health record-based chronic kidney disease registry to promote population health management. BMC Nephrol. 20, 1–11 (2019).
Norris, K. C. et al. Rationale and design of a multicenter Chronic Kidney Disease (CKD) and at-risk for CKD electronic health records-based registry: CURE-CKD. BMC Nephrol. 20, 1–9 (2019).
Navaneethan, S. D. et al. Development and validation of an electronic health record-based chronic kidney disease registry. Clin. J. Am. Soc. Nephrol. 6, 40–49 (2011).
Evans, K. et al. UK renal registry 20th annual report: introduction. Nephron 139, 1–11 (2018).
Pyart, R. et al. The 21st UK renal registry annual report: a summary of analyses of adult data in 2017. Nephron 144, 59–66 (2020).
Kramer, A. et al. The European Renal Association — European Dialysis and Transplant Association (ERA-EDTA) Registry Annual Report 2016: a summary. Clin. Kidney J. 12, 702–720 (2019).
McDonald, S. P. Australia and New Zealand dialysis and transplant registry. Kidney Int. Suppl. 5, 39–44 (2015).
Global Health Data Exchange. http://www.healthdata.org/about/ghdx (2020).
Rare Kidney Stone Consortium. http://www.rarekidneystones.org/ (2015).
Murdoch, T. B. & Detsky, A. S. The inevitable application of big data to health care. JAMA 309, 1351–1352 (2013).
Jensen, P. B., Jensen, L. J. & Brunak, S. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 13, 395–405 (2012).
McCartney, P. R. Clinical databases: electronic health records and repositories. MCN Am. J. Matern. Nurs. 38, 186 (2013).
Hripcsak, G. et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud. Health Technol. Inform. 216, 574–578 (2015).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 1–9 (2016).
Ta, C. N., Dumontier, M., Hripcsak, G., Tatonetti, N. P. & Weng, C. Columbia open health data, clinical concept prevalence and co-occurrence from electronic health records. Sci. Data 5, 1–17 (2018).
Centers for Medicare & Medicaid Services. CMS 2008-2010 Data Entrepreneurs’ Synthetic Public Use File (DE-SynPUF) (2019).
UK Biobank. Integrating Electronic Health Records into the UK Biobank Resource. http://biobank.ctsu.ox.ac.uk/showcase/showcase/docs/DataLinkageProcess.pdf (2014).
Visweswaran, S. et al. Accrual to clinical trials (ACT): a clinical and translational science award consortium network. JAMIA Open 1, 147–152 (2018).
The All of Us Research Program Investigators. The “All of Us” Research Program. N. Engl. J. Med. 381, 668–676 (2019).
Cadarette, S. M. & Wong, L. An introduction to health care administrative data. Can. J. Hosp. Pharm. 68, 232–237 (2015).
Nadkarni, G. N. et al. Development and validation of an electronic phenotyping algorithm for chronic kidney disease. AMIA Annu. Symp. Proc. 2014, 907–916 (2014).
Norton, J. M. et al. Development and validation of a pragmatic electronic phenotype for CKD. Clin. J. Am. Soc. Nephrol. 14, 1306–1314 (2019).
Wilkerson, M. L., Henricks, W. H., Castellani, W. J., Whitsitt, M. S. & Sinard, J. H. Management of laboratory data and information exchange in the electronic health record. Arch. Pathol. Lab. Med. 139, 319–327 (2015).
Mills, S. Electronic health records and use of clinical decision support. Crit. Care Nurs. Clin. North. Am. 31, 125–131 (2019).
Abdel-Kader, K. & Jhamb, M. EHR-based clinical trials: the next generation of evidence. Clin. J. Am. Soc. Nephrol. 15, 1050–1052 (2020).
Kohane, I. S. Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet. 12, 417–428 (2011).
Garcelon, N., Burgun, A., Salomon, R. & Neuraz, A. Electronic health records for the diagnosis of rare diseases. Kidney Int. 97, 676–686 (2020).
Matsushita, K. et al. Cohort profile: the chronic kidney disease prognosis consortium. Int. J. Epidemiol. 42, 1660–1668 (2013).
Tomašev, N. et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 572, 116–119 (2019).
Makino, M. et al. Artificial intelligence predicts the progression of diabetic kidney disease using big data machine learning. Sci. Rep. 9, 11862 (2019).
Akbilgic, O. et al. Machine learning to identify dialysis patients at high death risk. Kidney Int. Rep. 4, 1219–1229 (2019).
Ravizza, S. et al. Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat. Med. 25, 57–59 (2019).
Pivovarov, R., Albers, D. J., Sepulveda, J. L. & Elhadad, N. Identifying and mitigating biases in EHR laboratory tests. J. Biomed. Inform. 51, 24–34 (2014).
Sutton, P. R. & Payne, T. H. Interoperability of electronic health information and care of dialysis patients in the United States. Clin. J. Am. Soc. Nephrol. 14, 1536–1538 (2019).
Centers for Disease Control and Prevention. Surveillance Strategy Report — How Sharing Data Digitally Benefits Health. https://www.cdc.gov/surveillance/innovation/sharing-data-digitally.html (2018).
Krumholz, H. M. & Peterson, E. D. Open access to clinical trials data. JAMA 312, 1002–1003 (2014).
Baigent, C. et al. Challenges in conducting clinical trials in nephrology: conclusions from a Kidney Disease — Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 92, 297–305 (2017).
Kitchlu, A. et al. Representation of patients with chronic kidney disease in trials of cancer therapy. JAMA 319, 2437–2439 (2018).
Panchapakesan, U. & Pollock, C. Drug repurposing in kidney disease. Kidney Int. 94, 40–48 (2018).
Herrington, W. G., Staplin, N. & Haynes, R. Kidney disease trials for the 21st century: innovations in design and conduct. Nat. Rev. Nephrol. 16, 173–185 (2020).
Sim, I. et al. Time for NIH to lead on data sharing. Science 367, 1308–1309 (2020).
Kiley, R., Peatfield, T., Hansen, J. & Reddington, F. Data sharing from clinical trials — a research funder’s perspective. N. Engl. J. Med. 377, 1990–1992 (2017).
Mc Cord, K. A. et al. Routinely collected data for randomized trials: promises, barriers, and implications. Trials 19, 29 (2018).
Shlipak, M. & Stehman-Breen, C. Observational research databases in renal disease. J Am. Soc. Nephrol. 16, 3477–3484 (2005).
Loupy, A. et al. Prediction system for risk of allograft loss in patients receiving kidney transplants: International derivation and validation study. BMJ 366, l4923 (2019).
Egger, G. F. et al. European Union Clinical Trials Register: on the way to more transparency of clinical trial data. Expert Rev. Clin. Pharmacol. 6, 457–459 (2013).
Cochrane Kidney and Transplant. https://kidneyandtransplant.cochrane.org/ (2021).
Bierer, B. E., Li, R., Barnes, M. & Sim, I. A global, neutral platform for sharing trial data. N. Engl. J. Med. 374, 2411–2413 (2016).
Goldacre, B. & Gray, J. Opentrials: towards a collaborative open database of all available information on all clinical trials. Trials 17, 164 (2018).
Ross, J. S. et al. Overview and experience of the YODA project with clinical trial data sharing after 5 years. Sci. Data 5, 1–14 (2018).
Pencina, M. J. et al. Supporting open access to clinical trial data for researchers: the Duke Clinical Research Institute-Bristol-Myers Squibb supporting open access to researchers initiative. Am. Heart J. 172, 64–69 (2016).
Bhattacharya, S. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 5, 1–9 (2018).
Chen, J. et al. Assessment of postdonation outcomes in US living kidney donors using publicly available data sets. JAMA Netw. Open 2, e191851 (2019).
Sim, I. Mobile devices and health. N. Engl. J. Med. 381, 956–968 (2019).
Sieverdes, J. C. Mobile health considerations for kidney disease and transplantation. mHealth 4, 13–13 (2018).
Lambert, K., Mullan, J., Mansfield, K. & Owen, P. Should we recommend renal diet–related apps to our patients? An evaluation of the quality and health literacy demand of renal diet–related mobile applications. J. Ren. Nutr. 27, 430–438 (2017).
Streeper, N. M., Lehman, K. & Conroy, D. E. Acceptability of mobile health technology for promoting fluid consumption in patients with nephrolithiasis. Urology 122, 64–69 (2018).
Lunde, P., Nilsson, B. B., Bergland, A., Kværner, K. J. & Bye, A. The effectiveness of smartphone apps for lifestyle improvement in noncommunicable diseases: systematic review and meta-analyses. J. Med. Internet Res. 20, 1–12 (2018).
Singh, K. et al. Patients’ and nephrologists’ evaluation of patient-facing smartphone apps for CKD. Clin. J. Am. Soc. Nephrol. 14, 523–529 (2019).
Yang, Y., Chen, H., Qazi, H. & Morita, P. P. Intervention and evaluation of mobile health technologies in management of patients undergoing chronic dialysis: scoping review. JMIR mHealth Uhealth 8, e15549 (2020).
Pejchinovski, M. & Mischak, H. Clinical proteomics in kidney disease: from discovery to clinical application. Prilozi 38, 39–54 (2018).
Bullich, G. et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 94, 363–371 (2018).
Groopman, E. E., Rasouly, H. M. & Gharavi, A. G. Genomic medicine for kidney disease. Nat. Rev. Nephrol. 14, 83–104 (2018).
Groopman, E. E. et al. Diagnostic utility of exome sequencing for kidney disease. N. Engl. J. Med. 380, 142–151 (2019).
Weiss, R. H. Metabolomics and metabolic reprogramming in kidney cancer. Semin. Nephrol. 38, 175–182 (2018).
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol. 18, 1–15 (2017).
Papadopoulos, T. et al. Omics databases on kidney disease: where they can be found and how to benefit from them. Clin. Kidney J. 9, 343–352 (2016).
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A. & McKusick, V. A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, 514–517 (2005).
Lenffer, J. OMIA (Online Mendelian Inheritance in Animals): an enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res. 34, D599–D601 (2006).
Parsa, A. et al. Common variants in mendelian kidney disease genes and their association with renal function. J. Am. Soc. Nephrol. 24, 2105–2117 (2013).
Mallett, A. J. et al. Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int. 92, 1493–1506 (2017).
Tryka, K. A. et al. NCBI’s database of genotypes and phenotypes: DbGaP. Nucleic Acids Res. 42, 975–979 (2014).
Wong, K. M. et al. The dbGaP data browser: a new tool for browsing dbGaP controlled-access genomic data. Nucleic Acids Res. 45, D819–D826 (2017).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets — update. Nucleic Acids Res. 41, 991–995 (2013).
Papatheodorou, I. et al. Expression atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 46, D246–D251 (2018).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Thul, P. J. & Lindskog, C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 27, 233–244 (2018).
Yamamoto, T., Langham, R. G., Ronco, P., Knepper, M. A. & Thongboonkerd, V. Towards standard protocols and guidelines for urine proteomics: a report on the Human Kidney and Urine Proteome Project (HKUPP) symposium and workshop — 6 October 2007, Seoul, Korea and 1 November 2007, San Francisco, CA, USA. Proteomics 8, 2156–2159 (2008).
Shao, C. et al. A tool for biomarker discovery in the urinary proteome: a manually curated human and animal urine protein biomarker database. Mol. Cell. Proteom. 10, 1–8 (2011).
e-LICO An e-Laboratory for Interdisciplinary Collaborative Research in Data Mining and Data-Intensive Science. http://www.e-lico.eu/ (2019).
Jupp, S., Klein, J., Schanstra, J. & Stevens, R. Developing a kidney and urinary pathway knowledge base. J. Biomed. Semant. 2, S7 (2011).
Helfand, B. T., Mendez, M. G., Pugh, J., Delsert, C. & Goldman, R. D. Maintaining the shape of nerve cells. Mol. Biol. Cell 14, 5069–5081 (2003).
Chabardès-Garonne, D. et al. A panoramic view of gene expression in the human kidney. Proc. Natl Acad. Sci. USA 100, 13710–13715 (2003).
Willnow, T. E. et al. The European renal genome project. Organogenesis 2, 42–47 (2005).
Mischak, H. et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteom. Clin. Appl. 4, 464–478 (2010).
Moulos, P. et al. The KUPNetViz: a biological network viewer for multiple -omics datasets in kidney diseases. BMC Bioinformatics 14, 235 (2013).
Fernandes, M. & Husi, H. Establishment of a integrative multi-omics expression database CKDdb in the context of chronic kidney disease (CKD). Sci. Rep. 7, 1–11 (2017).
Zhao, H. et al. Kidney gene database: a curated and integrated database of genes involved in kidney disease. J. Urol. 172, 2344–2346 (2004).
Zhang, Q. et al. Renal Gene Expression Database (RGED): a relational database of gene expression profiles in kidney disease. Database 2014, 1–6 (2014).
Gillies, C. E. et al. An eQTL landscape of kidney tissue in human nephrotic syndrome. Am. J. Hum. Genet. 103, 232–244 (2018).
Qiu, C. et al. Renal compartment–specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 24, 1721–1731 (2018).
Ketchersid, T. Big data in nephrology: friend or foe? Blood Purif. 36, 160–164 (2014).
Shilo, S., Rossman, H. & Segal, E. Axes of a revolution: challenges and promises of big data in healthcare. Nat. Med. 26, 29–38 (2020).
Kaye, J. et al. Including all voices in international datasharing governance. Hum. Genomics 12, 18–23 (2018).
Reinholz, D. L. & Andrews, T. C. Breaking down silos working meeting: an approach to fostering cross-disciplinary STEM–DBER collaborations through working meetings. CBE Life Sci. Educ. 18, 1–8 (2019).
Davenport, T. & Kalakota, R. The potential for artificial intelligence in healthcare. Futur. Healthc. J. 6, 94–102 (2019).
Kruse, C. S., Goswamy, R., Raval, Y. & Marawi, S. Challenges and opportunities of big data in health care: a systematic review. JMIR Med. Inform. 4, e38 (2016).
Ngiam, K. Y. & Khor, I. W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 20, e262–e273 (2019).
Floege, J., Mak, R. H., Molitoris, B. A., Remuzzi, G. & Ronco, P. Nephrology research — the past, present and future. Nat. Rev. Nephrol. 11, 677–687 (2015).
Pépin, J. L., Bailly, S. & Tamisier, R. Big data in sleep apnoea: opportunities and challenges. Respirology 25, 486–494 (2019).
Adibuzzaman, M., DeLaurentis, P., Hill, J. & Benneyworth, B. D. Big data in healthcare — the promises, challenges and opportunities from a research perspective: a case study with a model database. AMIA Annu. Symp. Proc. 2017, 384–392 (2017).
Price, W. N. & Cohen, I. G. Privacy in the age of medical big data. Nat. Med. 25, 37–43 (2019).
Jeon, S. et al. Proposal and assessment of a de-identification strategy to enhance anonymity of the observational medical outcomes partnership common data model (OMOP-CDM) in a public cloud-computing environment: anonymization of medical data using privacy models. J. Med. Internet Res. 22, e19597 (2020).
Meskó, B. & Görög, M. A short guide for medical professionals in the era of artificial intelligence. NPJ Digit. Med. 3, 126 (2020).
Jha, A. K. et al. How common are electronic health records in the United States? A summary of the evidence. Health Aff. 25, 496–507 (2006).
Brennan, S. The biggest computer programme in the world ever! How’s it going? J. Inf. Technol. 22, 202–211 (2007).
Lee Ventola, C. Mobile devices and apps for health care professionals: uses and benefits. P T 39, 356–364 (2014).
Liu, C., Zhu, Q., Holroyd, K. A. & Seng, E. K. Status and trends of mobile-health applications for iOS devices: a developer’s perspective. J. Syst. Softw. 84, 2022–2033 (2011).
Sidey-Gibbons, J. A. M. & Sidey-Gibbons, C. J. Machine learning in medicine: a practical introduction. BMC Med. Res. Methodol. 19, 1–18 (2019).
Niel, O. & Bastard, P. Artificial intelligence in nephrology: core concepts, clinical applications, and perspectives. Am. J. Kidney Dis. 74, 803–810 (2019).
Geddes, C. C., Fox, J. G., Allison, M. E. M., Boulton-Jones, J. M. & Simpson, K. An artificial neural network can select patients at high risk of developing progressive IgA nephropathy more accurately than experienced nephrologists. Nephrol. Dial. Transplant. 13, 67–71 (1998).
Lin, K., Hu, Y. & Kong, G. Predicting in-hospital mortality of patients with acute kidney injury in the ICU using random forest model. Int. J. Med. Inform. 125, 55–61 (2019).
Gabutti, L. et al. Usefulness of artificial neural networks to predict follow-up dietary protein intake in hemodialysis patients. Kidney Int. 66, 399–407 (2004).
Akl, A. I., Sobh, M. A., Enab, Y. M. & James, T. Artificial intelligence: a new approach for prescription and monitoring of hemodialysis therapy. Am. J. Kidney Dis. 38, 1277–1283 (2001).
Barbieri, C. et al. An international observational study suggests that artificial intelligence for clinical decision support optimizes anemia management in hemodialysis patients. Kidney Int. 90, 422–442 (2016).
Acknowledgements
The authors would like to acknowledge Flavio Vincenti, Sri Lekha Tummalapalli, Vivek Rudrapatna, Douglas Arneson and Zicheng Hu (all University of California, San Francisco) for their valuable suggestions for this manuscript. The authors’ work was supported by the National Institute of Allergy and Infectious Diseases (Bioinformatics Support Contract HHSN316201200036W), the UCSF Bakar Computational Health Sciences Institute and the UCSF Clinical and Translational Sciences Institute, supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1 TR001872. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
N.K. researched data for the article and wrote the manuscript, S.B. contributed to data research and edited the manuscript. A.J.B. conceived the manuscript framework and reviewed and edited it before submission.
Corresponding author
Ethics declarations
Competing interests
A.T.B. is a co-founder and consultant to Personalis and NuMedii; consultant to Samsung, Mango Tree Corporation, and in the recent past, 10x Genomics, Helix, Pathway Genomics, and Verinata (Illumina); has served on paid advisory panels or boards for Geisinger Health, Regenstrief Institute, Gerson Lehman Group, AlphaSights, Covance, Novartis, Genentech, Merck and Roche; is a shareholder in Personalis and NuMedii; is a minor shareholder in Apple, Facebook, Alphabet (Google), Microsoft, Amazon, Snap, 10x Genomics, Illumina, CVS, Nuna Health, Assay Depot, Vet24seven, Regeneron, Sanofi, Royalty Pharma, AstraZeneca, Moderna, Biogen, Paraxel and Sutro, and several other non-health-related companies and mutual funds; and has received honoraria and travel reimbursement for invited talks from Johnson and Johnson, Roche, Genentech, Pfizer, Merck, Lilly, Takeda, Varian, Mars, Siemens, Optum, Abbott, Celgene, AstraZeneca, AbbVie, Westat, and many academic institutions, medical or disease-specific foundations and associations, and health systems. A.T.B. receives royalty payments through Stanford University, for several patents and other disclosures licensed to NuMedii and Personalis. His research has been funded by NIH, Northrup Grumman (as the prime on an NIH contract), Genentech, Johnson and Johnson, FDA, Robert Wood Johnson Foundation, Leon Lowenstein Foundation, Intervalien Foundation, Priscilla Chan and Mark Zuckerberg, the Barbara and Gerson Bakar Foundation, and in the recent past, the March of Dimes, Juvenile Diabetes Research Foundation, California Governor’s Office of Planning and Research, California Institute for Regenerative Medicine, L’Oreal, and Progenity.
Additional information
Peer review information
Nature Reviews Nephrology thanks Luxia Zhang, who co-reviewed with Chao Yang, William Herrington, Min Jun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Autosomal dominant polycystic kidney disease mutation database: https://pkdb.mayo.edu/
ClinicalStudyDataRequest.com: https://clinicalstudydatarequest.com/Default.aspx
Clinicaltrials.gov: https://clinicaltrials.gov/
Immune Tolerance Network: https://www.immunetolerance.org/
National Kidney Foundation Patient Network: https://www.kidney.org/nkfpatientnetwork
Nephroseq: https://www.nephroseq.org/resource/login.html
PatientsLikeMe: https://www.patientslikeme.com/
Registry Of Kidney Diseases: https://gardn.org.au/registries/registry-of-kidney-diseases/
RenDER data extraction and referencing system: https://render.usrds.org/render/xrender.phtml
Sentinel and Patient-Centered Outcomes Research Network: https://www.sentinelinitiative.org/sentinel/data/distributed-database-common-data-model
The HCUP National Inpatient Sample (NIS): https://www.hcup-us.ahrq.gov/nisoverview.jsp
The NephCure Kidney Network Patient Registry: https://nephcure.org/2015/12/the-nephcure-kidney-network-patient-registry-nkn/
Think Kidneys: https://www.thinkkidneys.nhs.uk/
WHO International Clinical Trials Registry Platform: https://www.who.int/ictrp/en/
Supplementary information
Glossary
- Deep learning
-
A type of machine learning that uses multiple layers to progressively extract higher level features from the input layer of the model. Common deep learning algorithms include convolutional neural networks, recurrent neural networks, general adversarial networks and autoencoders.
- k-anonymity
-
A data anonymization technique that protects the identities of individuals using methods such as suppression and generalization. A dataset is said to have k-anonymity if the information for each individual cannot be distinguished from that of at least k-1 individuals.
- l-diversity
-
A data anonymization approach that relies on introducing further entropy or diversity to the dataset. This model uses generalization and promotes diversity for sensitive values within a group. l-diversity is an extension of the k-anonymity model.
- t-closeness
-
This model is a further refinement of the k-anonymity and l-diversity models. t-closeness is the maximum of the distances between the distribution of values of a sensitive attribute and that of the entire database table. An equivalence class will have t-closeness if the distance between the attribute in the class and whole table is no more than threshold t.
Rights and permissions
About this article
Cite this article
Kaur, N., Bhattacharya, S. & Butte, A.J. Big Data in Nephrology. Nat Rev Nephrol 17, 676–687 (2021). https://doi.org/10.1038/s41581-021-00439-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41581-021-00439-x
This article is cited by
-
Blockchain in nephrology
Nature Reviews Nephrology (2023)
-
Predicting Renal Toxicity of Compounds with Deep Learning and Machine Learning Methods
SN Computer Science (2023)