Abstract
Disclosing the full potential of functional nanomaterials requires the optimization of synthetic protocols and an effective size screening tool, aiming at triggering their size-dependent properties. Here we demonstrate the successful combination of a wide-angle X-ray total scattering approach with a deep learning classifier for quantum dots sizing in both colloidal and dry states. This work offers a compelling alternative to the lengthy process of deriving sizing curves from transmission electron microscopy coupled with spectroscopic measurements, especially in the ultra-small size regime, where empirical functions exhibit larger discrepancies. The core of our algorithm is an all-convolutional neural network trained on Debye scattering equation simulations, incorporating atomistic models to capture structural and morphological features, and augmented with physics-informed perturbations to account for different predictable experimental conditions. The model performances are evaluated using both wide-angle X-ray total scattering simulations and experimental datasets collected on lead sulfide quantum dots, resulting in size classification accuracies surpassing 97%. With the developed deep learning size classifier, we overcome the need for calibration curves for quantum dots sizing and thanks to the unified modeling approach at the basis of the total scattering method implemented, we include simultaneously structural and microstructural aspects in the classification process. This algorithm can be complemented by incorporating input information from other experimental observations (e.g., small angle X-ray scattering data) and, after proper training with the pertinent simulations, can be extended to other classes of quantum dots, providing the nanoscience community with a powerful and broad tool to accelerate the development of functional (nano)materials.
Similar content being viewed by others
Introduction
The development of fast and reliable sizing methods for colloidal quantum dots (QDs) is of paramount importance to fully exploit their size-tunable optoelectronic properties1,2,3,4. In this regard, there is a general consensus on the use of the so-called “sizing curves”, which plays a crucial role in optimizing synthetic protocols and investigating size-dependent properties5. Typically, these sizing curves are empirical polynomials6,7,8,9 or inverse size-dependent terms added to the bulk bandgap5,10,11,12,13, mostly describing the trend of photophysical properties with size. They establish phenomenological relations between the average sizes of QDs, usually determined by transmission electron microscopy (TEM), and their absorption band edge energy, whose values are determined by the quantum confinement effect, which is particularly relevant for the size regimes investigated in syntheses and applications1. However, TEM has important critical issues, mainly related to inadequate statistics5 and underestimation of ultrasmall sizes, due to their low contrast with respect to the background signal14. Furthermore, TEM experiments are performed under vacuum and on dry QDs, whereas the corresponding spectroscopic data are collected on colloidal samples; this effect alone can potentially modify the sample by undesired ligand desorption and its size estimate if self-assembly phenomena trigger size-selection mechanisms15. Due to these intrinsic limitations of electron microscopy, small angle X-ray scattering (SAXS) has recently emerged as an alternative or complementary tool to define calibration curves for QDs sizing12,13,14. SAXS allows overcoming some of the TEM drawbacks, such as limitations in sample representativeness, and collecting data directly on colloidal suspensions. However, SAXS methods are not very routinary when it comes to data analysis, primarily due to the impact of QDs aggregation or self-assembly on SAXS data and to their sensitivity to (monodisperse) nanocrystals faceting, the reconstruction of which requires a precise description of fine morphological features of the sample16,17.
Despite their broad applicability to many QD categories, recent articles have drawn attention to significant discrepancies among empirical sizing expressions derived from different datasets for the same class of materials5,12,14. These differences have been attributed to either inaccurate size determination from TEM (related to the poor contrast between small particles and the underlying carbon support), or to unresolved band edge transitions (causing large errors in the first excitonic peak determination), and ultimately resulting in incomparable size estimation among different laboratories14.
A recent attempt to overcome the need for empirical calibration curves and to provide a physically meaningful tool for QDs sizing has been reported by Hens and coworkers12. They provide a semiempirical expression to describe quantum size effects for a large number of semiconductor nanocrystals based on the band gap of the bulk material, with a single fit parameter, employing a correction for the impact of nonparabolic energy bands on the QDs band gap. However, this approach, which considers quantum confinement as the sole origin of band gap tuning in QDs, has some limitations when structural aspects (e.g. lattice expansion/contraction, structural defects, octahedral rotations, etc.) play a role16,18,19,20.
Over the past decades, total scattering methods, in particular those based on the Debye Scattering Equation (DSE) and operating in reciprocal space, have been established as essential tools for characterizing the structure, microstructure and morphology of nanocrystals15,21,22,23, including ultrasmall QDs16,17,24. Although wide-angle scattering-based techniques are primarily sensitive to the atomic-scale structure of materials, reciprocal space total scattering methods provide robust information on multiple length scales, in particular if nanocrystalline materials are considered25. These goals are achieved by combining a data-collection strategy favoring high-angular resolution in Q-space26 (where Q = 4π sinθ/λ is the magnitude of the scattering vector), a strict data-reduction protocol and a unified structural and morphological modeling approach, starting from atomistic models and allowing the DSE to account for both Bragg and diffuse scatterings on equal footing27. This is particularly relevant for ultrasmall and/or defective QDs for which the peak broadening and diffuse scattering induced by size effects cannot be easily separated from those arising from structural defects. In such cases, a comprehensive modeling approach that incorporates all these features into atomistic models becomes necessary to extract quantitative and accurate structural and microstructural information17.
Nevertheless, constructing reliable and customized atomistic models, to be optimized against the experimental data to extract structural and microstructural parameters, remains a highly challenging task and often poses a bottleneck for wide-angle scattering-based methods28,29,30. To overcome this limitation, several semi-automated “mining” approaches have been recently developed, within the framework of real-space total scattering methods28,31,32,33,34,35,36,37,38. Inspired by these studies, we tackle the challenge of developing reliable, efficient, and user-friendly methods for QDs sizing, using a combination of reciprocal space wide-angle X-ray scattering methods based on the DSE and a convolutional neural network (CNN) that provides physically interpretable results.
In this work, we develop and apply this tool to lead-chalcogenide binary QDs, which serves as a benchmark system. Indeed they have been extensively characterized within the DSE approach, which has provided a well-established knowledge about their structural and morphological features16,24. Moreover, a notable aspect of these materials is the absence of planar defects, a high density of which typically challenges the determination of nanocrystal sizes by using wide-angle scattering methods17,19,21. The proposed supervised deep learning (DL) approach enables direct sizing of colloidal QDs within 3–5 s on standard personal computers (details are given in the Methods section), without the use of calibration curves, and employing wide-angle X-ray scattering data, easily accessible via synchrotron and even laboratory measurements, as sole input information. The simulated data used to train the CNN are modified with physics-informed data augmentation, through the implementation of experimental artifacts that perturb the synthetic X-ray scattering patterns. Importantly, our algorithm does not require any prior information on the material or strict requirements on the collected Q-range, and datasets of both colloidal and dry samples can be used, without significant loss of accuracy even down to a minimum Qmax∼4 Å−1. Therefore, the presented automated tool can be readily used for real-time sizing of PbS QDs via wide-angle X-ray patterns, even from diluted colloidal suspensions, within the limitations of the Q-range and signal-to-noise ratio typically encountered in in-situ and in-operando diffraction experiments or from fast data acquisition using a conventional laboratory diffractometer. Additionally, it can serve as a rapid screening tool for the optimization of synthetic protocols. Furthermore, the proposed method can be easily extended to other classes of nanocrystals, allowing non-experts in crystallography and X-ray diffraction (XRD) to use the proposed automated workflow to generate the required DSE pattern libraries used to train the CNN classifier.
Results and discussion
Debye Scattering Equation (DSE) simulations for training and testing datasets
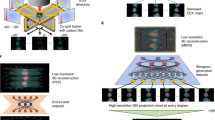
In this work, similar to other DL-based methods, we treated the X-ray scattering dataset as a holistic profile, analogous to an image. To train and test our model we first created a library of synthetic wide-angle X-ray total scattering (WAXTS) patterns computed by the DSE, following a bottom-up strategy, as illustrated in Fig. 1a (1. DSE simulations) and detailed in the Methods39. The use of simulated data, for the model training is motivated by practical considerations: as demonstrated elsewhere in a similar case of study30, the accuracy of the DL model is highly dependent on the size of the training set, and reaching a plateau requires a substantial amount of data. At this stage, an initial set of 1292 DSE simulations was created, by considering instrumental conditions that are rather standard for QDs studies at the Material Science beamline of the Swiss Light Source (0.5°–84° 2θ range, step 0.0036°, wavelength = 0.56 Å) and a combination of structural and microstructural parameters of lead sulfide (PbS) QDs, as follows: (i) for each size, 19 isotropic lattice deformation were included (from −0.25% to +0.51% of the PbS bulk lattice parameter 5.934(1) Å40, with a step of 0.04%), to account for size/ligands induced strain effects16; (ii) average diameters of PbS QDs from 2.0 to 10.0 nm (in steps of 0.5 nm), which encase the typical range of interest of strong quantum confinement regime for most common QDs12. This size range covers 17 classes, each of them combined with four different measures of size dispersion, in the form of standard deviations of a lognormal size distribution function (relative size dispersions of 5, 10, 15, 20%, considering the range of interest for QDs applications). Figure 1b showcases the effect of different lattice strains (−0.25% and +0.51% vs the bulk value, see Methods) and relative size dispersions (5% and 20%) on the DSE simulations of 3.0 nm and 8.0 nm PbS QDs.
a Schematics of the pipeline for the generation and standardization of the DSE X-ray pattern simulations to feed the CNN classifier. The standard workflow for training and testing the algorithm using DSE simulations is represented by blue arrows, while the red arrows show the path followed for additional testing (Robustness test) using newly created datasets. b DSE X-ray simulations of PbS QDs of average diameters of 3.0 nm (bottom line) and 8.0 nm (top line) generated with: relative lattice strain of +0.51% (red trace),−0.25% (blue trace) and 0% (green trace). The strain is computed as (a-aB)/aB × 100 (aB = 5.934(1) Å). Residual traces at the bottom correspond to −0.25%–0% and +0.51%–0% pattern differences; relative size dispersions of 5% (blue trace) and 20% (red trace). The size dispersions of the population of spherical nanocrystals, modeled as standard deviations of lognormal size distribution functions, are shown in the insets as histograms along with the number fraction of each cluster in the population; QDs concentrations in toluene (w/w = 7%-magenta, 20%-green, 44%-blue, 100%-red traces) used for training the CNN; signal to noise ratio (SNR) equal to 14 (red trace) and 122 (blue trace), which follows a Poisson distribution around the simulated values26.
Physics-informed DSE datasets augmentation
Typically, QDs are stored as colloidal suspensions of an organic solvent, and the WAXTS experiments can be performed either in capillaries filled with colloids or in dry conditions, by drop-casting or spin-coating the colloidal suspension on a flat substrate, or by letting them dry in open capillaries by solvent evaporation.
To bridge the gap between simulated patterns and real datasets, a physics-based data augmentation was implemented (2. Data Augmentation in Fig. 1a). This augmentation was obtained by combining each of the 1292 DSE simulations with (i) experimental WAXTS signals of two solvents commonly used for post-synthesis storage of QDs (hexane and toluene), properly rescaled to the PbS QDs trace to account for variations in the concentration of nanocrystals in the solvent. Four different QDs concentrations (7%, 20%, 44%, 100% in w/w% - where 100% indicates the dry condition) were treated. The corresponding DSE simulations are shown in Fig. 1b; (ii) four noise levels, chosen to represent the average signal/noise ratios (SNR) of 14, 24, 46, and 122 (Fig. 1b), according to a Poisson distribution26. By applying this augmentation method, 41344 WAXTS patterns were generated, resulting in an expanded dataset that encompasses a wide range of experimental conditions.
Before feeding the DSE patterns into the developed CNN, a data standardization step was implemented, corresponding to point 3. Data Standardization in Fig. 1a, to ensure a constant number of equispaced points, and a standardized integral area underlying the simulations. This pre-processing stage is aimed at obtaining uniformly scaled input datasets for both the training and the testing workflows shown in Fig. 1. The data standardization is applied to all input datasets, including simulated X-ray patterns and experimental data, and involves the following operations: (i) the x-scale of the X-ray patterns is converted from 2θ to Q, being \(Q=4\pi \sin {\rm{\theta }}/\lambda\) independent from the wavelength used for the experiments or simulations. This conversion is particularly convenient when dealing with synchrotron data, which can be collected with a wide range of different photon energies. (ii) A specific Q-range (1.0 Å−1 ≤ Q ≤ 15.0 Å−1) is selected from the input data. This approach eliminates the need for manual intervention to specify a range of the input vector for the DL model. If the Q-range of the input data falls outside the default choice of 1.0 Å−1 ≤ Q ≤ 15.0 Å−1, it is automatically adjusted: if longer, it is cut to fit the default range, if shorter, missing intensities are padded by adding a constant value obtained by averaging the last five intensity points of the trace (to mitigate fluctuations due to the noise). Therefore, the pre-selected Q values allow a multitude of different experimental ranges to be accommodated. (iii) The input pattern is sampled, through a spline interpolation, to have a final number of 5004 equispaced points in the selected Q-range (1.0 Å−1 ≤ Q ≤ 15.0 Å−1), with a Q-step of 0.0028 Å−1 (~0.016° in 2θ degrees). This step is particularly convenient when dealing with synchrotron data which are collected with high angular resolution and stored as several thousands of data points. iv) Each calculated pattern is rescaled to have the same integral area, in order to deal with comparable signals, which may significantly vary depending on the experimental conditions.
According to this strategy, a 1D vector with dimensions 5004 × 1 × 1 is generated from each DSE simulation (intensities only) of the database; these vectors are all collected in a comprehensive matrix used as input for the CNN.
The architecture of the final CNN classifier, inspired by that reported in ref. 30, consists of an input layer, followed by five convolutional layers, a Global Average Pooling Layer (GAP), and an output layer, as detailed in the Methods and in the Supplementary Information (Supplementary Methods and Supplementary Fig. 1). The final model presented in this work was trained on (randomly picked) 80% of the total DSE simulations (10% of which represents the validation set), and the remaining 20% of the dataset was used for testing. The cross-entropy loss function and accuracy curves across epochs for both the training and validation sets are reported in Supplementary Fig. 2.
We initially trained and tested the CNN using a single solvent (either hexane or toluene), whereas we evaluated the overall performances on a combined dataset, consisting of simulations with both solvents added. The outcomes revealed a poor performance of the algorithm under the aforementioned conditions, with accuracies as low as 60% (details are given in Supplementary Table 1). This result suggests a crucial influence of the type of solvent in accurately determining the size of particles in suspension, as highlighted in Supplementary Fig. 3. Therefore, we built the final expanded dataset by incorporating DSE traces that combine PbS QDs simulations with both toluene and hexane traces (41344 patterns). These are indeed the solvents commonly used for storage of colloidal suspensions of nanosized oleate-capped II-VI and IV-VI semiconductors.
To assess the robustness of the developed DL model across experimental conditions that were not accounted for during training, additional datasets were built for testing (Robustness tests in Fig. 1a). In this regard, a portion of the DSE simulations generated at point 2 in Fig. 1a (27% of the physics-augmented database), is randomly picked up and coupled with variations in Qmax values (5 reduced ranges, down to 2.50 Å−1) and Q-step (20 subgroups), down to 0.056 Å−1 (corresponding to 251 points only in the DSE pattern); moreover, an extra subset is generated by encoding new solvent contributions (corresponding to 11 new QDs concentrations, down to 2 w/w%).
Convolutional Neural Network (CNN) as QDs size classifier
The CNN was trained and tested on the augmented dataset, containing 41344 simulated patterns, achieving accuracies exceeding 97%.
The confusion matrices of this run (Supplementary Fig. 4), resulting from the fivefold cross-validation (as detailed in the Methods and Supplementary Methods), were evaluated to visualize and summarize the performance of the model, together with the histograms reporting the error distribution throughout the 17 classes (Fig. 2a) and for the QDs concentrations (in w/w%) used for training/testing the model (Fig. 2b).
Error distribution among the different size classes (a) and as a function of colloid dilution (b) cumulative for all sizes. Errors are calculated as the fraction of misclassifications during the testing phase with respect to the total number of predictions. This analysis provides insights into how the misclassifications are distributed across the 17 size classes (a) and colloid dilutions (b). c DSE simulations of PbS QDs with different average sizes, highlighting a progressive increase in the similarity of spectral features as the average diameter increases from small to large QDs; the Clark distance metric (dCl, very efficiently discriminating changes due to size effects)49 is computed between representative peaks shown in (c) (1.5 Å−1 ≤ Q ≤ 2.5 Å−1) as \(\sqrt{\mathop{\sum }\nolimits_{i,=1}^{N}{\left[\left|({I}_{i}^{{small}}-{I}_{i}^{{large}})\right|/({I}_{i}^{{small}}+{I}_{i}^{{large}})\right]}^{2}}\), Ii being the intensity at Qi for the two adjacent sizes. The smaller the dCl distance, the more similar the signals. The same trend is obtained by comparing the simulated full patterns (1.0 Å−1 ≤ Q ≤ 15.0 Å−1).
The size prediction accuracy (computed as the ratio of correct size classifications to the total number of predictions) decreases toward larger QD sizes, as shown in the confusion matrices of Supplementary Fig. 4. This result is further clarified by the analysis of the labels incorrectly assigned during testing, as illustrated in Fig. 2 and Supplementary Fig. 5, which provide a physical interpretation of the results obtained from the DL model. The number of errors of the CNN classifier rises with increasing PbS diameter (Fig. 2a), rather than being concentrated at the smaller sizes, for which the more significant peak broadening and solvent contribution (especially in highly diluted conditions) smear out the main diffraction peak features (Fig. 2c). In contrast, for the larger sizes, the Bragg peaks are quite sharp and well-defined, but more similar among adjacent classes (Fig. 2c). This observation emphasizes the limits in the applicability of the method, and in general of WAXS techniques, when dealing with larger average sizes, especially when coupled with low angular resolution instrumental setups due to the dominance of instrumental over sample features on the diffraction peaks26. On the other hand, an important implication of these results is the ability of the developed DL classifier to discriminate ultrasmall sizes (above reasonable dilution/SNR), typically one of the most challenging tasks at the nanoscale14.
To further analyze the effect of colloids dilution on QDs size classification, we evaluate the DL model by sorting the classified traces in the four QDs concentrations used for data augmentation (w/w = 7%, 20%, 44%, 100%), each concentration encompassing all sizes (Fig. 2b and Supplementary Fig. 5).
Very promising results are gained for the dry condition (w/w = 100%) for which no errors are found in size predictions. For the other concentrations, the number of misclassified elements positively correlates with the colloid dilution. This is quite an encouraging result, considering that the QDs colloidal suspensions can be easily drop-casted or spin-coated on a flat substrate, allowing the measurements to be performed in dry conditions using the typical lab XRD instrument in Bragg-Brentano geometry. It should be noted that (pseudo)dry conditions can always be obtained (even for QD colloidal suspensions) by subtracting the solvent scattering signal from the total scattering of the sample collected under the same experimental conditions. This is indeed a very convenient workaround, especially when dealing with solvents other than those used to train the CNN (toluene and hexane).
QDs size classification versus CNN regression model
To compare the performance on the same task, we also developed an alternative regression model for size prediction using a tailored CNN architecture (described in the Supplementary Methods) and the same database of 41344 augmented DSE simulations for training, validation, and testing. Supplementary Fig. 6 shows the mean square error loss function over epochs for both the training and validation (10% of the original training set) sets, along with the results of the predictions generated by this alternative model. The number of errors (calculated as the number of predictions deviating more than ± 0.25 nm from the corresponding true values, in line with the bin size of 0.50 nm used for size classification) is approximately twice that of the size classifier (accuracy ∼91%). This result is attributed to the intrinsic discrete nature of QDs well reproduced by the proposed atomistic model (see details in the Method section), which makes the size classifier more suitable than the regression model to the scope of determining the QDs size from WAXS data. On the other hand, a comparable error distribution to the classifier is found over sizes (Supplementary Fig. 6) and colloids concentration (Supplementary Fig. 7) when using the regression model, highlighting that this feature is primarily due to the information encoded in the input data rather than to the applied model.
Evaluating the robustness of the size classifier
We assessed the performance of the developed model in predicting the size of QDs while dealing with experimental artifacts different from those accounted for in the training set. To perform these robustness tests on the formerly trained model, we created additional datasets by varying the Qmax, Q-step, and QDs concentrations independently, within limits that were considered experimentally reasonable.
The developed model was fed with the new DSE simulations without retraining, and the accuracy in the classification of the PbS QDs size, defined as the ratio of correct size classifications to the total number of predictions, was computed as a function of the experimental parameters explored.
Firstly, we considered the robustness against the variation of the Q-range of the pattern simulations, here adjusted as variable Qmax depending on the experimental setup. Data collected either with laboratory instruments (typical Qmax ∼ 7 Å−1) or at synchrotron facilities with tunable Qmax (typically up to 12–15 Å−1 in high-angular resolution configurations41) are therefore considered. To this aim, we explored reduced ranges of that originally used for training the model (Qmin = 1.0 Å−1 and Qmax = 15.0 Å−1), by selecting five representative values in between. Figure 3a displays different values of Qmax each exemplified by color-coded vertical dashed lines, with the corresponding accuracy, in terms of model prediction, reported in Fig. 3b. In contrast to what is observed with real space total scattering methods36, applying Qmax truncation to reciprocal space data does not lead to irreparable signal distortions across the entire data range, instead, it results in a limited subset of information, contingent upon the explored range.
Q-range modification: (a) simulated DSE X-ray pattern of 3.0 nm PbS colloidal QDs in toluene, the dashed vertical lines represent the Qmax cut considered to estimate the robustness of the trained model. b Accuracy for size classification of PbS colloidal QDs, at different Qmax and the same Qmin = 1.0 Å−1. The colors of the markers correspond to the dotted vertical lines in (a), indicating the associated Qmax. Datasets coarsening: c simulated DSE X-ray patterns (y-offset for clarity) of representative 8.5 nm PbS colloidal QDs in toluene, and data coarsening from bottom (yellow trace 5004 points, Q-step = 0.0028 Å−1) to top (dark red trace 251 points, Q-step = 0.056 Å−1). A reduced Q-range is shown in the figure for the sake of clarity. d Accuracy for size classification of PbS colloidal QDs (x-axis in logscale) upon data coarsening (different Q-steps) at two representative Q-ranges: top 1.0 Å−1 ≤ Q ≤ 5.0 Å−1 (typical XRD lab. conditions, green markers); bottom: 1.0 Å−1 ≤ Q ≤ 15.0 Å−1 (typical synchrotron wide-angle X-ray scattering setup). QDs dilution: e simulated DSE patterns of 3.0 nm PbS colloidal QDs in toluene, characterized by different colloids concentrations, from w/w = 100% (black trace) down to w/w = 2% (red trace). A reduced Q-range is shown in the figure for the sake of clarity. f Accuracy for size classification of PbS colloidal QDs, considering the different colloid concentrations indicated on the x-axis. The colors of the markers correspond to the DSE simulations shown in (e).
Figure 3b shows that the accuracy of our model classifications remains consistently above 97% until a remarkably low Qmax value of 4.4 Å−1 is reached, a particularly relevant result considering limited experimental conditions, such as those accessible for example during in-situ/in-operando experiments. Below this threshold, a significant decay in the model’s performance is observed, with the accuracy dropping to 48% at Qmax = 2.5 Å−1, emphasizing the critical role of the low-Q data in accomplishing the targeted task. Indeed Qmin variation (tested at Qmin = 1.7 Å−1 and Qmax = 15.0 Å−1, not shown in Fig. 3 for the sake of clarity) produces an effect even if low-Q Bragg peaks are included in the simulations causing a substantial drop in the performances of the model down to 83% (Supplementary Fig. 8). It is noteworthy that the observed decrease in accuracy is somewhat dependent on the padding strategy employed: as reported in Supplementary Fig. 9, when a zero-filling approach is used to compensate for missing intensities within the reference Q-range, the accuracy of size predictions associated with Qmin = 1.7 Å−1 and Qmax = 15.0 Å−1 drops to 41%. In addition, slightly worse performances are associated with the same Qmax cutoffs shown in Fig. 3, probably due to significant changes in the overall scale of the X-ray scattering traces as a result of integrating the area over smaller Q-ranges. However, this effect is significantly mitigated by the padding strategy presented in Fig. 3, where the missing intensities are replaced by the value averaged over the last five intensity points, thus ensuring substantial invariance of the integrated pattern area. The error distribution analysis for the selected Q-ranges of Fig. 3 is reported in Supplementary Fig. 8, showing a higher accumulation of misclassifications at larger QDs sizes down to Qmax = 4.4 Å−1. The drops in accuracy observed for the Q-ranges 1.0–2.5 Å−1 and 1.7–15.0 Å−1 are accompanied by a general increase in the number of errors across all classes, indicating that the limitations imposed by these specific Q-ranges have a significant impact on the accuracy of the size classifier.
To further evaluate the tradeoff between accuracy and XRD data collection times, we investigated the impact of dataset coarsening on QDs sizing using the trained model (Fig. 3c, d and Supplementary Fig. 10). In Fig. 3c, we report representative DSE simulations of 8.5 nm colloidal QDs (10% of relative size dispersion) with increased Q-step from the bottom to the top.
Data coarsening was achieved by downsampling the original physics-augmented data used for training from 5004 points (Q-step = 0.0028 Å−1) down to 251 points (5% of the original number of points, Q-step = 0.056 Å−1), while preserving the Q-range. The accuracy in the size prediction was found to gradually decrease as the input data was coarsened, which corresponds to a decrease in both the number of observations and angular resolution. Remarkably, accuracies higher than 90% were preserved even when reducing the data points down to 25% of the original quantity in the range 1.0 Å−1 ≤ Q ≤ 15.0 Å−1 (Fig. 3b bottom, 1251 points, Q-step = 0.011 Å−1), and when data coarsening is applied to the reduced 1.0 Å−1 ≤ Q ≤ 5.0 Å−1 range for dried PbS QDs (Fig. 3b top, green markers), corresponding to an angular resolution of ∼0.2° with a Cu(Kα) X-ray source and mimicking the conditions of a typical XRD laboratory experiment. The latter result is of prime relevance in view of a massive application of this size classifier to XRD laboratory data, well matching the conditions of this robustness test.
The corresponding error analysis (Supplementary Fig. 10) indicates a more pronounced error accumulation at PbS sizes larger than 5 nm, while increasing the 2θ/Q-step of the input data. This observation can be attributed to the limited number of points in the coarsened dataset, which results in the smearing of the input information, more effective in the presence of sharper Bragg peaks, as it is for larger QDs sizes.
Reducing PbS QDs concentrations in the hexane/toluene colloidal suspensions, simulated by the decreasing of the scales ratio of PbS/solvent data in Fig. 3e, has a significant impact on the prediction performance, due to the smearing of the information content of the XRD traces, at constant Q-range (1.0 Å−1 ≤ Q ≤ 15.0 Å−1) and number of points (5004). Interestingly, similar to the Q-range truncation effect, the developed model preserves its ability to predict QDs size with an accuracy exceeding 90% across a wide range of concentrations (Fig. 3f), including values different from those used in the training set. It remains accurate down to concentrations as low as w/w = 6%, which is close to the minimum value employed in the training set (w/w = 7%). A larger number of errors starts to accumulate towards smaller sizes when colloid concentrations lower than 6% are reached (Supplementary Fig. 11). This result is because the size features encoded in the Bragg peaks width and shape become particularly blurred at low PbS/solvent ratios, as highlighted in Fig. 3e for 3 nm QDs size, thus increasing the uncertainty in size prediction for smaller QDs.
QDs sizing from experimental wide-angle X-ray total scattering (WAXTS) data
To validate the applicability of the developed size classifier to real cases, we applied it to QDs direct sizing by using experimental scattering data as sole input information. Experimental synchrotron data of PbS QDs in the 3–7 nm size range (Fig. 4a) were collected as colloidal suspensions in hexane or toluene and underwent the appropriate data reduction process described in the Methods section. Accurate DSE-based modeling was performed to extract detailed structural and microstructural information, as reported elsewhere16,24. It should be noted that the synchrotron setup used for this data collection ensures an adequate angular resolution, at the expense of a more limited Q-range (Qmax ∼15–17 Å−1). Nevertheless, when dealing with larger nanocrystal sizes (exceeding∼20–30 nm, out of the scope of this work) the potential impact of additional instrumental broadening on the WAXS data must be carefully evaluated26.
a WAXTS synchrotron data of PbS colloidal QDs in toluene (magenta trace) with different average diameters (as estimated by DSE data modeling reported in ref. 16). b Laboratory XRD data of PbS QDs deposited and dried on a monocrystalline silicon zero-background plate. c A comparison between DSE-estimated (DDSE) and CNN-predicted (DCNN) PbS colloidal QDs average diameters, showing an almost perfect linear correlation with y ∼ x. d PbS empirical sizing curves (purple, red, and blue dotted lines)14,42,43, and the semiempirical equation by Hens and coworkers reported in ref. 12 (gray solid line), showing the relationship between the first excitonic peak energy and the QDs diameter. The green squares represent the PbS reference diameters (D) determined through data fitting with the DSE (x-error bars are the e.s.d.’s of the associated lognormal size distribution functions, measuring the size dispersions), while the blue circles are the size predictions obtained with the DL size classifier developed in this work.
Additional laboratory XRD data have been collected in flat plate sample geometry, upon drop-casting of as-synthesized QDs and after a few months of aging. Figure 4b showcases a selection of these datasets. It is worth noting the much more restricted accessible Q-range than in synchrotron data, and that the peak intensities ratio of laboratory XRD data may differ significantly from those obtained in colloidal suspensions shown in Fig. 4a. This discrepancy is attributed to preferred orientation effects (texture) that occur when faceted QDs are deposited and dried on a flat substrate16. Our previous studies have shown that PbS QDs exhibit preferential alignment on {110} facets in fresh samples and {111}-{100} upon aging16, due to a progressive morphological evolution from a rhombic dodecahedral to a cuboctahedral shape. This less-than-perfect spherical morphology of QDs has important consequences when collecting XRD data from flat samples, resulting in altered peak intensity ratios, and partially hampering a thorough data analysis. This case further highlights the need for a modeless tool that can effectively extract size information from such type of data.
At this aim, we tested the size classifier developed in this work, on both texture-free synchrotron and textured laboratory data, without additional training (Fig. 4c and Supplementary Tables 2, 3). Very small deviations between the predicted and the reference average sizes (determined through DSE data fittings) are found. These discrepancies are estimated on average at 0.11 nm for synchrotron (Supplementary Table 2) and 0.25 nm for laboratory XRD data (Supplementary Table 3), both consistently less than 0.50 nm, that is the step size employed during the model training, representing indeed the limiting factor in the prediction performance. It is worth mentioning that employing a finer step size could potentially exceed the resolution of our method in the present case of study, particularly when coupled with the narrow-size dispersions typically exhibited by colloidal QDs, prepared with tailored syntheses. This limitation arises from the size discretization limit of the atomistic model construction database, which is defined by the diameter of the sphere having a volume equivalent to a single PbS primitive unit cell (0.46 nm, as detailed in the Methods).
The slightly worse performance of the size classifier when dealing with laboratory XRD data can be mostly attributed to the preferred orientation effects, which partially alter the intensity distributions “seen” by the model. Nevertheless, the method still exhibits very good accuracy even under these varied conditions. The accurate prediction of QD size dealing with both synchrotron and laboratory experimental data is highlighted in Fig. 4c in which the reference DSE sizes vs. the CNN classifier predictions are reported, showing a very good linear correlation at y∼x. Once again, worse performances are obtained with the regression model when applied to both synchrotron and laboratory experimental data, as outlined in Supplementary Fig. 12 and Supplementary Tables 4, 5, likely attributed to the limited size distribution of the samples analyzed16, emphasizing their inherent size discretization features favoring a more ‘rigid’ classification model, rather than a regression.
Figure 4d illustrates the relationship between the 1st excitonic peak energy and the diameter of PbS QDs using various empirical sizing curves, from TEM/SAXS (Tisdale and coworkers, red dashed line)42, SAXS (Hens and coworkers, purple dashed line)14, and WAXS (Ozin and coworkers, blue dashed line)43, some of them sourced from ref. 5. The gray solid line in Fig. 4d represents the generalized semiempirical sizing function recently proposed by Hens and coworkers12, which incorporates a correction for nonparabolic energy bands on the QDs band gap. This sizing curve for PbS QDs has been reproduced by using 45 nm as Bohr diameter, 17.4 for the (high frequency) dielectric constant, and 0.42 eV as optical band gap, as reported in ref. 12. Reference sizes (green squares)16,24, as well as predictions generated by our CNN classifier (blue circles) are also displayed. As noted in ref. 5, the empirical sizing curves for PbS exhibit the largest discrepancies at smaller QD sizes, likely stemming from the strong confinement regime in which lead chalcogenides reside. For this reason, PbS QDs demonstrate a more pronounced dependence of the band gap on their size compared to other binary IV-VI QDs5, emphasizing the potential for sizing errors and highlighting the need for robust methods for accurately determining the ultrasmall sizes of PbS QDs. On the other hand, our experimental results, derived from DSE data modeling and the CNN classifier presented in this work, nicely match the semiempirical sizing function derived by Hens and coworkers12 which takes into account precise physico-chemical considerations12. We further highlight the excellent match with the empirical curve by Ozin and coworkers from XRD data and conventional size analysis based on the approximated Scherrer equation, suggesting that peak broadening in PbS QDs originates mainly from finite-size effects, and it is not affected by structural defects.
In this work, we have developed a DL model that enables fast, accurate, and fully automated sizing of QDs by using wide-angle X-ray scattering data as sole input information, and without the need for calibration curves, paving the way for alternative AI-based methods for nanocrystals characterization.
We have addressed several experimental challenges by implementing a physically meaningful data augmentation, which enhances the flexibility of the model to handle extreme conditions encountered in experiments, such as low QDs concentrations and SNR for colloidal suspensions, resulting from rapid data collection or limited material availability.
We have shown that the proposed approach exhibits excellent performance (accuracy exceeding 90%) even under untrained experimental conditions such as a very limited Q-range of the input data (Qmax ∼4 Å−1), coarsening of the dataset (down to a Q-step of ~ 0.01 Å−1), and reduced colloidal QDs concentrations (w/w > 6%). This suggests that reasonably high accuracies can be maintained with significantly reduced XRD data collection times and/or angular resolutions, particularly for smaller QDs.
The validity of our approach, which combines a CNN classifier with reciprocal space X-ray scattering methods, is strongly supported by the excellent agreement observed between our size classifications and those obtained by accurate DSE data modeling and validated by TEM analysis16. Moreover, our PbS QDs size predictions align perfectly with the generalized semiempirical sizing function recently proposed by Hens and coworkers12 which includes a correction for non-parabolic bands on the QDs band gap.
We would like to emphasize that the methodology proposed here is intended as a simplified and reliable tool for conducting fast-size screening for QDs in the 2–10 nm range, particularly in situations requiring a fast response (e.g. during the optimization of synthetic methods); additionally, our model can be easily integrated into high-throughput experimental workflows, including in-situ/in-operando experiments, even by non-experts in crystallography and XRD. Moreover, this pioneering integration of reciprocal space X-ray total scattering and DL, which allows direct sizing of QDs both in colloidal and dry states, addresses some of the limitations of traditional methods based on empirical calibration curves, which are inherently limited in their general applicability and often hampered by the different experimental conditions required for TEM (dry samples) and optical spectroscopy (colloidal QDs). By combining the detailed multiscale information, from atomic-to-the nanometer length scales, accessible through the DSE-based approach with the performance predicting capabilities of DL, we intend to promote an original perspective in functional (nano)material characterization.
The approach proposed here can be extended to predict structural and microstructural properties of different classes of QDs and semiconductor nanocrystals from XRD and total scattering data, provided that appropriate training is performed. The training process can be easily facilitated using the comprehensive set of tools developed in this study, which complements the fast DSE computation from atomistic models of nanocrystals, already available through the distributed Debussy Suite of programs (https://debyeusersystem.github.io)39.
Further developments in this field, e.g., addressing the sizing of nanocrystals characterized by anisotropic morphologies (for which the different growth directions are not accessible by TEM characterization) or other atomic precise information (like lattice strains), and complementing the scattering information with other experimental and computational methods, are envisaged in the near future.
Methods
Debye Scattering Equation (DSE) simulations from atomistic models of PbS QDs
The DSE provides the average differential cross-section (or the powder diffraction pattern) of a randomly oriented powder from the distribution of interatomic distances between atomic pairs, without any assumption of periodicity and order27,44:
where Q = 4πsinθ/λ is the magnitude of the scattering vector, λ is the radiation wavelength, fi is the atomic form factor of element i, dij is the interatomic distance between atoms i and j, N is the total number of atoms and T and o are the thermal atomic displacement parameter and the site occupancy factor associated to each atomic species, respectively. The first summation in the above equation includes the contributions of zero distances between one atom and itself and the second term (the interference term) the non-zero interatomic distances dij = |ri − rj|.
The DSE-based simulations in the present work were performed using the DebUsSy Suite of programs39, relying on a two-step approach. In the first step, starting from the structural information encoded in the Crystallographic Information File for the bulk material40, we generated atomistic models PbS QDs of spherical shape and increasing size. To create each cluster of the database, we generated a lattice of nodes and dressed it with a rhombohedral unit cell, that is the primitive unit cell corresponding to the face-centered cubic structure reported for the bulk material (cell edge of the primitive cell a = 4.196 Å vs ak = 5.934(1) Å in the fcc structure)40. This choice is motivated by the advantage of reducing the step size between adjacent clusters in the population, thus ensuring an increased resolution in terms of size retrieval16. Accordingly, the final monovariate population of spherical PbS QDs contains 45 clusters in the size range 0.46–20.87 nm with a constant step of 0.46 nm, which corresponds to the diameter of a sphere of volume equivalent to one PbS primitive unit cell.
Gaussian sampled interatomic distances45 and related pseudo-multiplicities are calculated from the atomistic models of PbS QDs and encoded in suitable databases.
The DSE equation is computed in the second step by using the structural and microstructural information detailed in the main text and fed by the sets of sampled interatomic distances calculated in the first step.
Convolutional Neural Network (CNN) architecture
The architecture of the CNN for size classification developed in this work is detailed in the Supplementary Information (Supplementary Methods and Supplementary Fig. 1). Briefly, it consists of an input layer (1D input vector with dimensions 5004 × 1 × 1), followed by five convolutional layers, with 32 kernels each, and strides/kernel sizes of 10, 5, 4, 3, and 2 units respectively. After the final convolutional layer, a flattening step is performed by using a Global Average Pooling Layer (GAP), which replaces conventional fully connected layers. The use of a GAP offers advantages in terms of reinforcing the direct correspondence between feature maps and classes thereby promoting a more physically interpretable classification. Additionally, it helps mitigate the risk of overfitting46. The final output layer of the model consists of 17 nodes, representing the 17 classes of QDs sizes for classification.
This CNN was trained on 80% of the 41344 total simulated data, and the remaining 20% of the dataset was used for testing. To address potential overfitting issues, 10% of the training set was used as validation set. Fivefold cross-validation was performed, to check whether different parts of the dataset lead to different performances (Supplementary Fig. 4), and the cross-entropy loss function and model accuracy for both the training and validation sets were monitored across epochs (Supplementary Fig. 2).
The whole training/validation/testing process of the network was performed in 10 min on a GPU-enabled personal computer (Intel Core i7-12700H processor; NVIDIA GeForce RTX 3060 graphics card) and in about 35 min on a multi-core processor (AMD EPYC 7301 16-Core processor). Once the model was developed, the size prediction took 3–5 s, as tested on the same PCs used for the training and on an Apple Macbook Air with a 1.7 GHz Intel Core i7 processor.
Details on the architecture of the developed neural network implementing a regression method are reported in the Supplementary Information.
Wide-Angle X-ray Total Scattering (WAXTS) data collection and reduction
To evaluate the performance of the developed deep learning (DL) model in predicting the average sizes of PbS quantum dots (QDs), we used experimental datasets from a series of synchrotron WAXTS experiments. These experiments were conducted directly on colloidal suspensions of fourteen PbS QDs samples ranging from 3 to 8 nm in size, dispersed in hexane or toluene, inside borosilicate glass capillaries of 0.7–0.8 mm in diameter. The data collection was performed at the X04SA-MS Powder diffraction beamline of the Swiss Light Source (SLS, PSI)47, using a position-sensitive single-photon counting 1D-detector (MYTHEN-II)48. Two different beam energies of 25 KeV and 22 KeV were used, and the corresponding operational wavelengths were precisely determined by measuring a silicon powder standard (NIST 640d, a0 = 0.543123(8) nm at 22.5 °C) under the same experimental conditions. All datasets were collected in the 0.5–120° 2θ-range with a step of 0.0036°. Independent scattering curves were obtained for air and empty capillaries, as well as empty and sample-loaded direct beam transmissions. These additional measurements were necessary to perform angle-dependent absorption corrections and subtract any extra-sample scattering contributions. Inelastic Compton scattering is added in the DSE simulations as an additional component.
X-ray powder diffraction (XRD) measurements
Laboratory XRD data were collected and analyzed as described in ref. 16 on dried samples. A droplet of each colloidal sample was deposited on the surface of a silicon monocrystal zero-background plate with the aid of a micropipette and dried in air within minutes. The XRD diffractograms were collected using Cu-Kα radiation (λ = 1.5418 Å) on a Bruker AXS D8 Advance Diffractometer equipped with a Lynxeye detector operating at 40 kV and 40 mA. Occasionally, data were also collected on a Rigaku Miniflex diffractometer equipped with a DTEX detector operating at 30 kV and 10 mA. No significant differences were observed between the two instrumental setups, as contributions of the instrumental broadening to peak shapes and widths were negligible in both cases. The measured angular ranges for all datasets are characterized by a 2θmin = 20° and a 2θmax ranging between 80° and 120°, with a common 2θ-step of 0.02°.
Data availability
The training and test simulations used in this work are publicly available through https://github.com/DeByeUSerSYstem/QDots-sizer. All other data are available from the corresponding authors on reasonable request.
Code availability
All codes developed and implemented in this work can be found in a public repository located at https://github.com/DeByeUSerSYstem/QDots-sizer.
References
Kovalenko, M. V. et al. Prospects of nanoscience with nanocrystals. ACS Nano 9, 1012–1057 (2015).
Carey, G. H. et al. Colloidal quantum dot solar cells. Chem. Rev. 115, 12732–12763 (2015).
de Mello Donega, C. Synthesis and properties of colloidal heteronanocrystals. Chem. Soc. Rev. 40, 1512–1546 (2011).
Zhang, J. et al. Colloidal quantum dots: synthesis, composition, structure, and emerging optoelectronic applications. Laser Photonics Rev. 17, 2200551 (2023).
Kuno, M., Gushchina, I., Toso, S. & Trepalin, V. No one size fits all: semiconductor nanocrystal sizing curves. J. Phys. Chem. C. 126, 11867–11874 (2022).
Jasieniak, J., Smith, L., van Embden, J., Mulvaney, P. & Califano, M. Re-examination of the size-dependent absorption properties of CdSe. Quantum Dots. J. Phys. Chem. C. 113, 19468–19474 (2009).
de Mello Donegá, C. & Koole, R. Size dependence of the spontaneous emission rate and absorption cross section of CdSe and CdTe quantum dots. J. Phys. Chem. C 113, 6511–6520 (2009).
Lin, S. et al. Surface and intrinsic contributions to extinction properties of ZnSe quantum dots. Nano Res. 13, 824–831 (2020).
Moreels, I. et al. Composition and size-dependent extinction coefficient of colloidal PbSe quantum dots. Chem. Mater. 19, 6101–6106 (2007).
Moreels, I. et al. Size-dependent optical properties of colloidal PbS quantum dots. ACS Nano 3, 3023–3030 (2009).
Capek, R. K. et al. Optical properties of zincblende cadmium selenide quantum dots. J. Phys. Chem. C 114, 6371–6376 (2010).
Aubert, T. et al. General expression for the size-dependent optical properties of quantum dots. Nano Lett. 22, 1778–1785 (2022).
Toufanian, R., Zhong, X., Kays, J. C., Saeboe, A. M. & Dennis, A. M. Correlating ZnSe quantum dot absorption with particle size and concentration. Chem. Mater. 33, 7527–7536 (2021).
Maes, J. et al. Size and concentration determination of colloidal nanocrystals by small-angle X-ray scattering. Chem. Mater. 30, 3952–3962 (2018).
Bertolotti, F. et al. Size segregation and atomic structural coherence in spontaneous assemblies of colloidal cesium lead halide nanocrystals. Chem. Mater. 34, 594–608 (2022).
Bertolotti, F. et al. Crystal symmetry breaking and vacancies in colloidal lead chalcogenide quantum dots. Nat. Mater. 15, 987–994 (2016).
Moscheni, D. et al. Size-dependent fault-driven relaxation and faceting in zincblende CdSe colloidal quantum dots. ACS Nano 12, 12558–12570 (2018).
Prasanna, R. et al. Band gap tuning via lattice contraction and octahedral tilting in perovskite materials for photovoltaics. J. Am. Chem. Soc. 139, 11117–11124 (2017).
Bertolotti, F. et al. Band gap narrowing in silane-grafted ZnO nanocrystals. A comprehensive study by wide-angle X-ray total scattering methods. J. Phys. Chem. C 125, 4806–4819 (2021).
Frison, R. et al. Magnetite–Maghemite nanoparticles in the 5–15 nm range: correlating the core–shell composition and the surface structure to the magnetic properties. A total scattering study. Chem. Mater. 25, 4820–4827 (2013).
Bertolotti, F. et al. A total scattering Debye function analysis study of faulted Pt nanocrystals embedded in a porous matrix. Acta Crystallogr. A 72, 632–644 (2016).
Bertolotti, F. et al. Coherent nanotwins and dynamic disorder in cesium lead Halide Perovskite nanocrystals. ACS Nano 11, 3819–3831 (2017).
Bertolotti, F. et al. Crystal structure, morphology, and surface termination of Cyan-Emissive, six-monolayers-thick CsPbBr3 nanoplatelets from X-ray total scattering. ACS Nano 13, 14294–14307 (2019).
Bertolotti, F. et al. Ligand-induced symmetry breaking, size and morphology in colloidal lead sulfide QDs: from classic to thiourea precursors. Chem. Sq. 2, 1–14 (2018).
Ferri, F., Bertolotti, F., Guagliardi, A. & Masciocchi, N. Nanoparticle size distribution from inversion of wide angle X-ray total scattering data. Sci. Rep. 10, 12759 (2020).
Dengo, N., Masciocchi, N., Cervellino, A., Guagliardi, A. & Bertolotti, F. Effects of structural and microstructural features on the total scattering pattern of nanocrystalline materials. Nanomaterials 12, 1252 (2022).
Bertolotti, F., Moscheni, D., Guagliardi, A. & Masciocchi, N. When crystals go nano - the role of advanced x-ray total scattering methods in nanotechnology. Eur. J. Inorg. Chem. 2018, 3789–3803 (2018).
Anker, A. S. et al. Extracting structural motifs from pair distribution function data of nanostructures using explainable machine learning. npj Comput. Mater. 8, 1–11 (2022).
Szymanski, N. J., Bartel, C. J., Zeng, Y., Tu, Q. & Ceder, G. Probabilistic deep learning approach to automate the interpretation of multi-phase diffraction spectra. Chem. Mater. 33, 4204–4215 (2021).
Oviedo, F. et al. Fast and interpretable classification of small X-ray diffraction datasets using data augmentation and deep neural networks. npj Comput. Mater. 5, 1–9 (2019).
Banerjee, S. et al. Cluster-mining: an approach for determining core structures of metallic nanoparticles from atomic pair distribution function data. Acta Cryst. A 76, 24–31 (2020).
Yang, L., Juhás, P., Terban, M. W., Tucker, M. G. & Billinge, S. J. L. Structure-mining: screening structure models by automated fitting to the atomic pair distribution function over large numbers of models. Acta Cryst. A 76, 395–409 (2020).
Magnard, N. P. L., Anker, A. S., Aalling-Frederiksen, O., Kirsch, A. & Jensen, K. M. Ø. Characterisation of intergrowth in metal oxide materials using structure-mining: the case of γ-MnO2. Dalton Trans. 51, 17150–17161 (2022).
Kjær, E. T. S. et al. In situ studies of the formation of tungsten and niobium oxide nanoparticles: towards automated analysis of reaction pathways from PDF analysis using the Pearson correlation coefficient. Chem.–Methods 2, e202200034 (2022).
Liu, C.-H., Tao, Y., Hsu, D., Du, Q. & Billinge, S. J. L. Using a machine learning approach to determine the space group of a structure from the atomic pair distribution function. Acta Cryst. A 75, 633–643 (2019).
Lan, L., Liu, C.-H., Du, Q. & Billinge, S. J. L. Robustness test of the spacegroupMining model for determining space groups from atomic pair distribution function data. J. Appl. Cryst. 55, 626–630 (2022).
Kjær, E. T. S. et al. DeepStruc: towards structure solution from pair distribution function data using deep generative models. Digit. Discov. 2, 69–80 (2023).
Anker, A. S. et al. Characterising the atomic structure of mono-metallic nanoparticles from X-ray scattering data using conditional generative models. Preprint at https://doi.org/10.26434/chemrxiv.12662222.v1 (2020).
Cervellino, A., Frison, R., Bertolotti, F. & Guagliardi, A. DEBUSSY 2.0: the new release of a Debye user system for nanocrystalline and/or disordered materials. J. Appl. Cryst. 48, 2026–2032 (2015).
Noda, Y., Ohba, S., Sato, S. & Saito, Y. Charge distribution and atomic thermal vibration in lead chalcogenide crystals. Acta Cryst. B 39, 312–317 (1983).
Chupas, P. J. et al. Rapid-acquisition pair distribution function (RA-PDF) analysis. J. Appl. Cryst. 36, 1342–1347 (2003).
Weidman, M. C., Beck, M. E., Hoffman, R. S., Prins, F. & Tisdale, W. A. Monodisperse, air-stable PbS nanocrystals via precursor stoichiometry control. ACS Nano 8, 6363–6371 (2014).
Cademartiri, L. et al. Size-dependent extinction coefficients of PbS quantum dots. J. Am. Chem. Soc. 128, 10337–10346 (2006).
Debye, P. Zerstreuung von Röntgenstrahlen. Ann. Phys. 351, 809–823 (1915).
Cervellino, A., Giannini, C. & Guagliardi, A. On the efficient evaluation of Fourier patterns for nanoparticles and clusters. J. Comput. Chem. 27, 995–1008 (2006).
Lin, M., Chen, Q. & Yan, S. Network in network. Preprint at https://arxiv.org/abs/1312.4400v3 (2013).
Willmott, P. R. et al. The materials science beamline upgrade at the Swiss Light Source. J. Synchrotron Radiat. 20, 667–682 (2013).
Bergamaschi, A. et al. The MYTHEN detector for X-ray powder diffraction experiments at the Swiss Light Source. J. Synchrotron. Radiat. 17, 653–668 (2010).
Hernández-Rivera, E., Coleman, S. P. & Tschopp, M. A. Using similarity metrics to quantify differences in high-throughput data sets: application to X-ray diffraction patterns. ACS Comb. Sci. 19, 25–36 (2017).
Acknowledgements
A.G. contributed in the framework of the research activities carried out within the Project “Network 4 Energy Sustainable Transition—NEST”, Spoke 1., Project code PE0000021, funded under the National Recovery and Resilience Plan (NRRP), Mission 4, Component 2, Investment 1.3— Call for tender No. 1561 of 11.10.2022 of Ministero dell’Universita‘ e della Ricerca (MUR); funded by the European Union—NextGenerationEU. F.B. acknowledges Fondazione Cariplo, Project nr: 2020-4382 - CubaGREEN for partial financial support.
Author information
Authors and Affiliations
Contributions
A.G. and F.B. conceived the research, collected and modeled the synchrotron WAXTS and laboratory XRD data from PbS QDs. L.A. developed, implemented, tested the CNN model, and processed the results, with key intellectual contributions from A.G. and F.B.; A.G. and F.B. wrote the manuscript, with input from L.A.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Allara, L., Bertolotti, F. & Guagliardi, A. A deep learning approach for quantum dots sizing from wide-angle X-ray scattering data. npj Comput Mater 10, 54 (2024). https://doi.org/10.1038/s41524-024-01241-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-024-01241-6