Abstract
Melanoma cells are highly resistant to conventional genotoxic agents, and BRAFV600/MEK-targeted therapies as well as immunotherapies frequently remain inefficient. Alternative means to treat melanoma, in particular through the induction of programmed cell death modalities such as apoptosis or necroptosis, therefore still need to be explored. Here, we report that melanoma cell lines expressing notable amounts of RIPK1, RIPK3 and MLKL, the key players of necroptosis signal transduction, fail to execute necroptotic cell death. Interestingly, the activity of transforming growth factor β-activated kinase 1 (TAK1) appears to prevent RIPK1 from contributing to cell death induction, since TAK1 inhibition by (5Z)-7-Oxozeaenol, deletion of MAP3K7 or the expression of inactive TAK1 were sufficient to sensitize melanoma cells to RIPK1-dependent cell death in response to TNFα or TRAIL based combination treatments. However, cell death was executed exclusively by apoptosis, even when RIPK3 expression was high. In addition, TAK1 inhibitor (5Z)-7-Oxozeaenol suppressed intrinsic or treatment-induced pro-survival signaling as well as the secretion of cytokines and soluble factors associated with melanoma disease progression. Correspondingly, elevated expression of TAK1 correlates with reduced disease free survival in patients diagnosed with primary melanoma. Overall, our results therefore demonstrate that TAK1 suppresses the susceptibility to RIPK1-dependent cell death and that high expression of TAK1 indicates an increased risk for disease progression in melanoma.
Similar content being viewed by others
Introduction
Melanoma are highly chemoresistant and, despite the success of BRAF/MEK-targeting therapeutics and immune checkpoint inhibitor-based treatments, many patients still do not benefit from these novel treatment options or experience disease relapse [1, 2]. An attractive addition to the armament against malignant melanoma could be treatments that induce programmed cell death through receptors of the tumor necrosis factor (TNF) family. TNF receptor activation can trigger two signaling cascades towards cell death, namely apoptosis and necroptosis [3, 4]. Apoptosis relies on the FADD-dependent activation of caspase-8 at the death inducing signaling complex, the subsequent permeabilisation of the outer mitochondrial membrane and the activation of the apoptosis execution network [5]. Necroptosis likewise is FADD-dependent in this context but, in contrast to protease-driven apoptosis, is transduced by a kinase signaling cascade in which activating (auto-)phosphorylation of RIPK1, RIPK3 and MLKL culminates in the latter forming lytic pores in the plasma membrane [6, 7]. Apoptosis and necroptosis signaling branches exert interplay, with caspase-8 proteolytically inactivating RIPK1 and RIPK3, and RIPK1 modulating apoptosis and necroptosis susceptibility depending on its activity, phosphorylation, and ubiquitylation status [3, 8, 9].
If not inducing cell death, the activation of TNF receptors and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptors stimulates pro-survival signaling [3, 4]. For example, tumor-intrinsic TNFα contributes to resistance against BRAF(V600) inhibitor-based treatments and suppresses tumor-directed immune responses [10, 11]. This highlights the need to combine TNF receptor ligands with therapeutics that reliably direct signaling towards programmed cell death. A major component of the pro-survival signaling complexes that form prior to apoptosis- or necroptosis-inducing platforms are cellular inhibitor of apoptosis proteins (cIAPs) 1 and 2. cIAPs recruit the linear ubiquitin chain-assembly complex (LUBAC), that is essential to activate the canonical NFκB pathway and that limits caspase-8 and RIPK1 recruitment [4, 12]. High affinity IAP-antagonists are therefore attractive co-treatments to enhance programmed cell death in response to TNFα or TRAIL [13]. Besides cIAPs, also transforming growth factor β-activated kinase 1 (TAK1) has been implicated in regulating cell death susceptibility in this context. TAK1 contributes to pro-survival signaling by activating MAPK and NFκB pathways and additionally directly modulates apoptosis and necroptosis susceptibility by phosphorylating RIPK1 [14,15,16].
Previous work demonstrated that RIPK3-deficient melanoma cells understandably are necroptosis resistant [17]. Here, we therefore studied the cell death responsiveness and cell death modalities observed in melanoma cells that intrinsically express notable amounts of RIPK1, RIPK3 and MLKL. We likewise investigated the role of TAK1 in controlling cell death susceptibility and its potential as a prognostic marker for disease progression in melanoma.
Materials and methods
Reagents and antibodies
zVAD-fmk (Z-Val-Ala-Asp-fluoromethyl ketone) was purchased from Apexbio (MA, USA). The IAP antagonist compound A was generously provided by Tetralogics Corp. (Philadelphia, PA, USA). Necrostatin-1 was from Seleckchem (Houston, TX, USA). (5Z)-7-Oxozeaenol and MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) were obtained from Sigma-Aldrich (St. Louis, USA). NG-25 was purchased from MedChem Express (New Jersey, USA). Recombinant human TNF-α was purchased from R&D Systems (Minneapolis, MN, USA). Fc-scTRAIL (TRAIL) was kindly provided by Prof. Roland Kontermann (University of Stuttgart, Germany). The following antibodies were used: rabbit polyclonal antibody RIPK3 (Cat No: IMG-5846A, IMGENEX, San Diego, Ca, USA), mouse monoclonal RIPK1 (Cat. No: R41220, Transduction Laboratories, San Diego, California), mouse monoclonal PARP (BD556494), rabbit monoclonal phospho-MLKL (ab187091), rabbit monoclonal phospho-RIPK1 (CST-65746), rabbit monoclonal phospho-RIPK3 (ab209384), mouse monoclonal phospho-ERK1/2 (sc-7383), mouse monoclonal Caspase-8 (CST-9746), rabbit polyclonal Caspase-3 (CST-9662), rabbit monoclonal DNMT1 (CST-5032), mouse monoclonal Actin (CST-3700), rabbit monoclonal TAK1 (CST-4505), rabbit monoclonal phospho-TAK1 (CST-4580), rabbit monoclonal NF-ĸB p65 (CST-8242), rabbit monoclonal phospho-NF-ĸB (CST-3033), mouse monoclonal IĸBα (CST-4814), rabbit monoclonal phospho-IĸBα (CST-2859), mouse monoclonal phospho-p38 (CST-9216), rabbit polyclonal p38 (CST-9212), mouse monoclonal ERK1/2 (CST-9107). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit, goat anti-rat IgG, and goat anti-mouse IgG were obtained from Dianova GmbH (Hamburg, Germany)
Cell culture
The following human melanoma cell lines were obtained either from the ATCC (WM115, SK-MEL 1, SK-MEL 5, WM793B, Malme-3M, MeWo, and SK-MEL 2; Manassas, VA, USA), the Wistar Institute (WM35, WM852, WM3211, WM1366, WM1346 and WM1791C; Philadelphia, PA, USA), the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (MEL-JUSO; Brunswick, Germany), Rockland Immunochemicals Inc (WM852 and WM3248; Limerick, PA, USA), Memorial Sloan-Kettering Cancer Center (WM1346 and SK-MEL 147; New York, USA). Eight cell lines carried activating BRAF mutations (WM35, WM793B, WM852, WM3248, WM115, SK-MEL 1, SK-MEL 5, and Malme-3M), five cell lines NRAS mutations (WM1366, WM1346, SK-MEL 147, SK-MEL 2, MEL-JUSO), one cell line a CDK4 mutation (WM1791C), one cell line carried a c-KIT mutation (WM3211), and one cell line was BRAF/NRAS/c-KIT/CDK4 wildtype (MeWo). Cells were cultured in Roswell Park Memorial Institute medium (RPMI) with 10% fetal calf serum, at 5% CO2 at 37 °C. HaCaT keratinocytes were kindly provided by Petra Boukamp (DKFZ, Heidelberg, Germany) and cultured in RPMI medium as described above. Cell lines used in this study were tested regularly to be free of mycoplasm and were authenticated through STR profiles.
siRNAs, plasmids, and transfections
The following siRNA duplexes were used: FlexiTube siRNA for caspase-8 (#Hs_CASP8_11, Qiagen, Germany), for RIPK1 (#Hs_RIPK1_5, Qiagen, Germany), and the corresponding control siRNA (AllStars Negative Control siRNA 1027281, Qiagen, Germany). siRNA (10 nM) and Lipofectamine RNAiMAX (Invitrogen, CA, USA) were diluted in Opti-MEM medium and incubated for 5 min at room temperature prior to addition to cells. Procedures followed the manufacturer’s recommendations. Plasmids pCMV-TAK1-WT, pCMV-TAK1-K63W and empty vector control were kindly provided by Prof. Jun Ninomiya-Tsuji, NC State University, Raleigh, USA and described previously [18]. Cells were transfected by using ViaFectTM transfection reagent (Cat no: E4981; Promega Corporation, Madison, USA) according to the manufacturer’s instructions.
Generation of TAK1 deficient cells
TAK1 deficient WM1791C cells were generated by the CRISPR/Cas9 approach. Cells were transfected with empty vector pSpCas9(BB)-2A-GFP-PX458 or a vector targeting TAK1 (Cat no: 48138, Addgene, Cat no: sc-400364, Santa Cruz Biotechnology). Two days after transfection, GFP positive clones were isolated by cell sorting (BD FACSAriaTM III, BD Biosciences) and plated as single clones in 96-well plates. Clones were cultured for 2–3 weeks and analysed for successful TAK1 knockout by Western blotting.
MTT assay
For viability assays, 0.5–2 × 104 cells were seeded per well of a 96 well plate and incubated overnight at 37 °C. Prior to experimental endpoints, cells were incubated with MTT at 5 mg/ml for 3–4 h and kept maintained in cell culture incubators. Medium was then aspirated, 100 µl methanol were added, followed by 30 min incubation. The optical density of each well was measured with a TECAN multi reader (Infinite® 200 PRO series, Männedorf, Switzerland) at 570 nm (OD 570 nm). Cellular viability was calculated using untreated controls as reference samples.
Cell death assay
Cells were collected and stained with propidium iodide (2 μg/ml) for 15 min in the dark at room temperature. Flow cytometric measurements were performed using a MACSQuant Analyzer 10 (Miltenyi Biotech) and data were analysed using Flowing Software (Turku Centre for Biotechnology, Finland).
Western blotting
Cells were washed with ice-cold PBS and lysed for 1 h on ice in cell lysis buffer (30 mM TRIS-HCl (pH 7.5), 120 mM NaCl, 10% Glycerol, 1% Triton X, 1X Complete Protease Inhibitor (Roche, Mannheim, Germany). Cellular debris was removed by centrifugation at 20,000×g for 20 min. Protein concentrations were quantified by Bradford assay. Equal amounts of proteins were supplemented with 5 × Laemmli sample buffer and heated to 95 °C for 5 min. Proteins were separated on Nu-Page 4–12% Bis-Tris gels (Invitrogen, USA) and transferred to nitrocellulose membranes using an iBlot 2 gel transfer device (Invitrogen, Rockford, USA). After 1 h blocking with 5% bovine serum albumin (BSA) in Tris Buffered Saline with Tween-20 (TBST), the membranes were incubated with primary antibodies (diluted in 5% BSA) overnight. After washing with TBST, the blots were incubated with horseradish peroxidase (HRP)-conjugated isotype-specific anti-mouse and/or anti-rabbit secondary antibodies. After further washing, chemiluminescence signals were detected on film or using an AmershamTM imager 600 system (Amersham Bioscience Europe, Freiburg, Germany).
xMAP (luminex) assays
Cells were cultured in 96-well plates, grown to near confluence and treated as indicated. Supernatants were transferred to separate 96-well plates and cells were lysed with ProATonce lysis buffer (ProtATonce, Athens, Greece) by freezing/thawing. Cellular debris was removed by centrifugation at 2700×g for 20 min and the protein concentration of the lysates was adjusted to 250 µg/µl using a BCA assay kit (Pierce™). xMAP bead-based ELISA type assays were performed on a Luminex FLEXMAP 3D® platform (Luminex, Austin, TX), using a custom-developed phosphoprotein 3-plex panel (ProtATonce, Athens, Greece): Mitogen-activated protein kinase-3 (ERK1/MAPK3)—T202/Y204, Transcription factor AP-1 (c-JUN)—S63 and Transcription factor p65 (p65/Nf-κB)—S536. For cytokine measurements, the protein concentrations of supernatants were adjusted to 100 µg/µl and cytokines were measured using a custom-developed 4-plex panel (ProtATonce, Athens, Greece): Interleukin-8 (IL8), Intercellular adhesion molecule (ICAM1), Follistatin (FST), chemokine (C-C motif) ligand 5 (CCL5) and Chemokine (C-X-C motif) ligand 1 (CCXL1).
Survival analysis
For analysis of primary melanoma data stored in the cancer genome atlas (TCGA), normalized mRNA expression data (Upper Quartile normalized Fragments per Kilobase of transcript per Million mapped read, log2(FPKM-UQ+1)) generated by the Genomic Data Commons (GDC-NIH) were downloaded from the UCSC-XENA browser (UCSC Xena. Available at: https://xena.ucsc.edu/. (Accessed: 24 October 2018). The “disease free survival from sample procurement” was calculated as the difference between the “disease free survival from initial diagnosis” and “sample procurement from initial diagnosis”, downloaded from cBioPortal [19] and the GDC data portal (https://portal.gdc.cancer.gov/. (Accessed: 25 October 2018)), respectively. If no recurrence or progression events were reported, the date of last follow up or death were considered as final time points. An optimized chi-squared based separator was determined to divide patients with high and low MAP3K7 (TAK1) mRNA expression, and the two groups were compared by log-rank test. The optimal separator resulted in the highest chi-squared value with the function survdiff of the library survival in R (version 3.4.0), and the difference in mean survival time at different cut-off points was calculated and plotted with the tool Cutoff Finder [20]. Kaplan-Meier survival curves were generated and compared using GraphPad Prism 4.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7. If not stated otherwise, the mean values of three independently conducted experiments are plotted, where bars sh7ow the standard error of mean. Data were analyzed for statistically significant differences using One-way ANOVA and Bonferroni correction for multiple testing. A confidence level of 95% was used for all analyses (α-value = 0.05). Results were summarized as ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; n.s. = not significant.
Results
Melanoma cells expressing RIPK1, RIPK3 and MLKL are necroptosis resistant
Cancer cells frequently downregulate key necroptosis actors, in particular RIPK3 [17, 21, 22]. To study necroptosis competency, we therefore initially assessed expression heterogeneity of the essential RIPK1-RIPK3-MLKL necroptosis axis in melanoma cell lines. Across a panel of 16 cell lines, including cell lines harboring activating BRAF and NRAS mutations (see materials and methods), we noted comparable RIPK1 expression whereas RIPK3 and MLKL amounts differed considerably (Fig. 1a). RIPK3 was absent in many cell lines but frequently also clearly detectable, up to amounts similar to those observed in whole cell protein extracts from necroptosis competent HaCat keratinocytes [17]. With the exception of SK-MEL-2 and MeWo cells, all other cell lines expressed clearly detectable but differing amounts of MLKL (Fig. 1a). All cell lines expressed comparable amounts of procaspase-8 and cFLIP, as regulators of apoptosis signal transduction and, with exception of cIAP2, also comparable amounts of XIAP and cIAP1, as negative regulators of both apoptosis and necroptosis signaling [3] (Fig. 1b). To study necroptosis competency, we selected Malme-3M, MelJuso, WM1791C and WM3211 cells for subsequent experiments, since these cells expressed RIPK3 and MLKL, albeit in differing amounts.
Melanoma cells expressing RIPK1, RIPK3 and MLKL are necroptosis resistant. a, b Basal expression of key cell death regulatory proteins in melanoma cell lines was determined by immunoblotting. Necroptosis-competent HaCat keratinocytes served for comparison. Actin served as loading control. c Cell viability of MEL-JUSO, WM1791C, WM3211, and Malme-3M cell lines in response to TNFα, TRAIL or IAP antagonist Compound A, as determined by MTT assays. Cells were treated for 24 h. Data show mean ± S.E.M. from n = 3 independent experiments. d, e Cell viability in response to combination treatment of TNFα or TRAIL with IAP antagonist Compound A, as determined by MTT assays. Cells were treated for 48 h. zVAD-fmk and Necrostatin 1 were added 30 min prior to treatment. HT-29 cells served as necroptosis competent control. Data show mean ± S.E.M. from n = 3 independent experiments
TNF-receptor pathways can induce necroptosis, in particular when combining TNF receptor ligands with IAP antagonists and, to suppress apoptosis signaling, caspase inhibitors such as zVAD-fmk [23]. With the exception of moderately responsive MelJuso cells, all cell lines were largely resistant to TNFα or a multivalent TRAIL variant [24] (Fig. 1c). IAP antagonist Compound A reduced viability in Malme-3M cells, but only at very high concentrations (Fig. 1c). The combination of TNFα and Compound A most prominently reduced cell viability in WM3211 and Malme-3M cells, and the combination with caspase inhibitor zVAD-fmk (zVAD) failed to reduce viability further, indicating that canonical necroptosis cannot be induced (Fig. 1d). Correspondingly, RIPK1 inhibitor necrostatin-1 (nec-1) did not notably prevent viability loss in WM3211 and Malme-3M cells. WM1791C and MEL-JUSO cells likewise failed to die by necroptosis (Fig. 1d). Similar results were obtained in TRAIL-based treatment scenarios, where combination treatment with Compound A prominently reduced cell viability, and with zVAD but not nec-1 offering protection (Fig. 1d), indicating that cell death proceeds by apoptosis rather than necroptosis. Indeed, depleting caspase-8 expression in melanoma cells was sufficient to prevent cell death (Supplementary Fig. 1). In contrast, nec-1 or the combination of zVAD and nec-1 rescued necroptosis competent HT29 cells from TNFα/Compound A or TRAIL/Compound A treatment (Fig. 1e). Taken together, these data therefore demonstrate that melanoma cell lines with evident expression of the key actors of the canonical necroptosis cascade, namely RIPK1, RIPK3, MLKL, are incapable of dying by necroptosis.
Elevating intrinsic RIP3 expression fails to establish necroptosis competency in melanoma
It was previously demonstrated that DNA methyltransferase activity might silence RIPK3 expression [22, 25]. We therefore reasoned that treatment with 5-aza-2′-deoxycytidine (5-AD), a demethylating agent that induces the degradation of DNA (cytosine-5)-methyltransferase 1 (DNMT1) [26], could increase intrinsic RIPK3 expression and therefore necroptosis competency in melanoma cells. Indeed, 5-AD treatment notably enhanced RIPK3 expression in all cell lines, whereas RIPK1 expression remained unaffected (Fig. 2a). In MALME-3M and MEL-JUSO cells, also MLKL expression increased following 5-AD treatment (Fig. 2a). As expected, DNMT1 expression was lost in cells exposed to 5-AD (Fig. 2b). Importantly, RIPK3 and MLKL protein amounts remained elevated for several days after 5-AD withdrawal (Fig. 2c), indicating that 5-AD pre-treatment might pose an attractive strategy for priming cells towards RIPK3/MLKL-dependent necroptosis. However, despite substantially elevated RIPK3 expression, no substantial induction of necroptosis could be observed (Fig. 2d), indicating that signal transduction towards necroptosis is likely inhibited upstream of RIPK3/MLKL signaling.
Elevating intrinsic RIP3 expression fails to establish necroptosis competency in melanoma. a Melanoma cells were treated with 5-AD for 4 days and whole cell extracts were probed for RIPK3, RIPK1, MLKL expression amounts by immunoblotting. Actin served as loading control. b DNMT1 expression was determined following 2 days of 5-AD treatment by immunoblotting. Actin served as loading control. c Elevated RIPK3 expression persists after 5-AD withdrawal. As in (a), protein amounts were probed in melanoma cell populations up to 5 days after 5-AD washout. d Cell viability of melanoma cells in which intrinsic RIPK3 expression was elevated by 5-AD pre-treatment. Viability was determined by MTT assays. Data show mean ± S.E.M. from n = 3 independent experiments
TAK1 inhibitor (5Z)-7-Oxozeaenol establishes RIPK1-dependent apoptosis but not necroptosis competency in melanoma
Besides activating NFκB and MAPK signaling pathways, the transforming growth factor-β-activated kinase 1 (TAK1) has been implicated in suppressing RIPK1 activity and thereby to suppress necroptosis signaling [16]. We therefore reasoned that antagonizing TAK1 activity, using the high affinity inhibitor (5Z)-7-oxozeaenol (Oxo) [27, 28], might restore necroptosis competency in malignant melanoma. As expected, Oxo suppressed NFκB and MAPK signaling, as demonstrated by loss of phosphorylated p65 and IκBα (Fig. 3a) and by reductions in p38 and ERK1/2 phosphorylation (Fig. 3b). Oxo did not affect the expression of RIPK1, RIPK3 or MLKL (Fig. 3c).
(5Z)-7-Oxozeaenol establishes conditions of caspase- and RIPK1-dependent cell death in TNFα/Compound A-treated cells. a, b WM1791C cells were pre-treated for 30 min with 5z-7-oxozeaenol, followed by 30 min of TNF-α stimulation. Whole cell extracts were studied by immunoblotting. Actin served as loading control. c WM1791C cells were treated for 2 h and whole cell extracts were probed for RIPK1, RIPK3, and MLKL expression amounts by immunoblotting. Actin served as loading control. d–f Cell viability of melanoma cells or HT29 cells treated as indicated. Viability was determined by MTT assays following 24 h of treatment. Data show mean ± S.E.M. from n = 3 independent experiments. g WM 1791C cells were treated as indicated. Whole cell extracts were analyzed 2 h after stimulation with TNFα. Actin served as loading control
As a single drug or in combination with Compound A, Oxo was only modestly effective in reducing melanoma cell viability, whereas the triple combination with TNFα strongly reduced cellular viability in all cell lines tested (Fig. 3d). A similar effect was observed when using an alternative TAK-1 inhibitor (NG-25) (Fig. 3e). Viability loss in WM3211 was completely caspase dependent, whereas in WM1791C cells, and to a lesser extent in Malme-3M cells, RIPK1 inhibition by nec-1 prevented viability loss (Fig. 3d). In MEL-JUSO cells, nec-1 reduced viability loss only in combination with caspase-inhibitor zVAD (Fig. 3d). In contrast to necroptosis competent HT29 cells (Fig. 3f), Oxo therefore established conditions of caspase and RIPK1-dependent cell death in TNFα/Compound A-treated cells, as seen prominently in WM1791C cells. Interestingly, cell death remained completely independent of RIPK1 in TRAIL-based treatment scenarios (Supplementary Fig. 2).
In addition, Oxo notably suppressed TNFα and/or Compound A-induced pro-survival NFκB signaling, as demonstrated by reduced amounts of phosphorylated p65 and IκBα in Oxo-combination treatments (Fig. 3g). Luminex-based phosphoprotein measurements provided similar results, with Oxo preventing basal or TNFα/Compound A-induced phosphorylation of p65, Jun kinase and MAP kinase 3 (MAPK3) (Supplementary Fig. 3A). Oxo treatments also reduced basal or TNFα/Compound A-induced secretion of cytokines and soluble factors that were previously linked to melanoma disease progression and escape from immune surveillance, such as interleukin-8 (IL8) [29, 30], chemokine (C-X-C motif) ligand 1 (CXCL1) [31, 32], chemokine (C-C motif) ligand 5 (CCL5) [33], follistatin (FST) [34], and shedded intercellular adhesion molecule 1 (ICAM-1) [35] (Supplementary Fig. 3b).
Overall, these data therefore indicate that TAK-1 inhibitor Oxo enhances cell death susceptibility of melanoma cells exposed to TNFα/Compound A. To more specifically test for RIPK1 or caspase-8 dependency in this treatment scenario, we depleted RIPK1 and caspase-8 expression in WM1791C cells. In line with our previous results, viability loss required the presence of RIPK1 and caspase-8 (Fig. 4a). Processing of caspase-8 and 3 as well as PARP cleavage was prominently detectable after treatment with TNFα, Compound A and Oxo, with both zVAD and nec-1 suppressing this apoptotic cascade (Fig. 4b). Cell morphologies and cell death measurements by PI staining further supported that cell death was executed primarily by RIPK1 dependent apoptosis (Fig. 4c, d). Taken together, Oxo therefore appears to enhance the competency of melanoma cells to execute RIPK1 dependent apoptosis, but not necroptosis.
Cell death sensitization by (5Z)-7-Oxozeaenol requires the presence of RIPK1 and caspase-8. a Cell viability of WM1791C cells in which RIPK1 and/or caspase-8 expression were depleted by siRNA transfection (48 h), following 24 h of treatment. Data show mean ± S.E.M. from n = 3 independent experiments. Immunoblots demonstrate depletion efficiency. b WM1791C cells were treated as indicated and whole cell extracts were studied by immunoblotting. Actin served as loading control. c Cells were treated for 24 h as indicated and observed by phase contrast microscopy. d Cell death quantification for WM1791C cells treated for 24 h as indicated. Cell death was determined by flow cytometric measurement of propidium iodide positive cells. Data show means ± S.E.M. from n = 3 independent experiments
Necroptosis remains blocked upon combination of 5-AD pre-treatment and addition of (5Z)-7-oxozeaenol
Since Oxo treatment established conditions of RIPK1-dependent apoptosis in melanoma cells, we next tested if absence of necroptosis was attributable to the low expression of RIPK3 in melanoma. We therefore used melanoma cells pre-treated with 5-AD (see Fig. 2) to test if combination treatments with TNFα/Compound A/Oxo were now capable of inducing necroptotic cell death. As seen in Fig. 5a, none of the cell lines studied died by necroptotic cell death. However, the dependency on RIPK1 activity increased in WM3211 and WM1791C cells, as seen when combining zVAD and nec-1 (Fig. 5a), possibly due to elevated amounts of RIPK3 supporting RIPK1-dependent apoptosis in this setting [36]. To study how far necroptosis signaling is transduced through the RIPK1, RIPK3, MLKL cascade, we studied the phosphorylation status of these proteins. While the conventional necroptosis-inducing treatment of TNFα/Compound A/zVAD resulted in strong auto-phosphorylation of RIPK1 at Ser166 and in phosphorylation of RIPK3 (Ser227) and MLKL (Ser358) in HT-29 cells, not even RIPK1 was phosphorylated in WM1791C cells (Fig. 5b). This finding indicates early termination of necroptosis signaling in the latter cell line. In WM1791C cells pre-treated with 5-AD and exposed to treatment combinations as in Fig. 5a, RIPK1 auto-phosphorylation was observed in combination with the triple treatment of TNFα/Compound A/Oxo, but only when caspase activation was prevented (Fig. 5c). Caspase activity therefore seems to prevent RIPK1 oligomerisation, as needed for auto-phosphorylation and as seen in necroptosis competent HT-29 cells, and caspase inhibition was insufficient for signaling to proceed from RIPK1 towards RIPK3 and MLKL.
Necroptosis remains blocked upon combination treatment with 5-AD and (5Z)-7-oxozeaenol. a Cell lines in which RIPK3 expression was restored by 5-AD treatment (1 µM; 4d) were subsequently treated as indicated. Cell viability was measured by MTT assays. Data show means ± S.E.M. from n = 3 independent experiments. b HT-29 and WM1791C cells were treated for 24 h as indicated and whole cell extracts were probed for the amounts of phosphorylated or overall RIPK1, RIPK3 and MLKL. Actin served as loading control. c WM1791C cells were treated with 5-AD for 4d and afterwards treated for 24 h, as indicated. Whole cell extracts were probed for the amounts of phosphorylated or overall RIPK1, RIPK3 and MLKL. Actin served as loading control
TAK-1 inactivation is essential for RIPK1-dependent apoptosis in melanoma
Even though Oxo is a high affinity inhibitor of TAK1, off target effects cannot be excluded based on its selectivity profile [28]. We therefore performed genetic studies to test the relevance of TAK1 in preventing RIPK1-dependent apoptosis in melanoma more specifically. First, we eliminated TAK1 expression by CRISPR/Cas9 targeting of the MAP3K7 gene and studied viability loss after treatment with TNFα/Compound A (Fig. 6a, b). WM1791C cells lacking TAK1, but not parental control cells, were substantially sensitized to treatment with TNFα/Compound A, and cell death could be completely prevented by zVAD or nec-1 (Fig. 6b). Cell morphologies and propidium iodide uptake returned similar results, indicating apoptosis induction specifically for the TNFα/Compound A combination treatment in cells lacking TAK1 (Fig. 6c). We next restored TAK1 expression by introducing either wildtype or kinase dead TAK1 (Fig. 6d). Only wildtype TAK1 but not inactive TAK1 protected against the combination of TNFα/Compound A (Fig. 6e), demonstrating TAK1 and specifically its kinase activity to be a key suppressor of RIP1-dependent apoptosis in melanoma.
TAK-1 inactivation is essential for RIPK1-dependent apoptosis in melanoma. a Confirmation of TAK1 knock out in WM1719C cells by immunoblotting. Actin served as loading control. b Control and TAK1-deficient WM1719C cells were treated for 24 h as indicated. Cell viability was determined by MTT assay. Data show means ± S.E.M. from n = 3 independent experiments. c Cells were treated for 24 h as indicated and observed by phase contrast microscopy. Cell death was subsequently determined by flow cytometric measurement of propidium iodide positive cells. Data show means ± S.E.M. from n = 3 independent experiments. d Confirmation of TAK1 reconstitution (wild type or K63W) by immunoblotting, following transient transfection with respective expression vectors. e Control and TAK1-reconstituted cells were treated for 24 h as indicated. Cell viability was determined by MTT assay. Data show means ± S.E.M. from n = 3 independent experiments
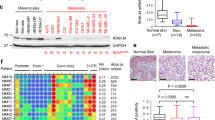
Since TNFα can be found within melanoma microenvironments [37], elevated expression of TAK1 in melanoma cells could contribute to preventing TNFα-induced apoptotic signaling and additionally might promote migration and invasion through other signaling pathways, as was shown in breast and colon cancer [38]. We therefore studied how the expression of MAP3K7 relates to disease progression in melanoma in a cohort of n = 102 patients diagnosed with primary melanoma for which dates of sample procurement and sufficient follow up information was available (Fig. 7a). The mean disease free survival between patients with low and high MAP3K7 expression in primary melanomas differed across a range of group separator thresholds (Fig. 7b), with significantly better prognosis for patients with low MAP3K7 expression (Fig. 7c). These results therefore extend our findings on the cell death-suppressing role of TAK1 in melanoma cell lines towards delineating a role of TAK1 in melanoma pathophysiology and patient outcome.
Low expression of TAK1 in primary melanoma samples is associated with favourable patient prognosis. a Cohort characteristics (n = 102 patients). b Difference in mean disease free survival (DFS) including the 95% confidence interval obtained by screening for a group separator for high and low MAP3K7 mRNA expression. The optimal separator is indicated by a vertical line. The distribution of normalized expression of MAP3K7 mRNA (log2(FPKM-UQ + 1)) is shown as a rug plot at the bottom of the graph. c Survival analysis in the SKCM-TCGA primary sub-cohort (n = 102 patients). Disease free survival (DFS) from sample procurement of patients with high (red) and low (blue) MAP3K7 mRNA expression was compared by log-rank test
Discussion
Here, we report that melanoma cells expressing the key players of necroptosis signal transduction fail to execute necroptotic cell death. However, signaling triggered by TNF receptor activation can be directed towards RIPK1-dependent apoptosis by inhibition of TAK1. Indeed, low expression of TAK1 is associated with a more favorable prognosis for patients diagnosed with primary melanoma.
Whereas RIPK1 was expressed homogeneously across the panel of melanoma cell lines studied here, we noted considerable heterogeneity in RIPK3 and MLKL expression. While primary melanocytes and nevus cells express RIPK3 [17], RIPK3 expression appears to be lost during melanomagenesis and is typically absent in most commonly used cell line models. Interestingly, the presence of BRAF and AXL oncogenes correlates with a loss in RIPK3 expression, as was recently shown in a large cell line panel [21]. In our melanoma cell line panel, the majority of BRAF mutated cells lines expressed low or undetectable amounts of RIPK3, with the exception of SK-MEL-1 and Malme-3M, both of which expressed very high amounts of RIPK3. How BRAF or AXL activity suppresses the expression of RIPK3 and how relevant and broadly applicable such a mechanism is in the melanoma setting, however, will need further investigation. We showed that silencing of RIPK3 expression in melanoma likely depends on DNA methylation, since 5-AD treatment and the associated degradation of DNMT1 correlated with the accumulation of RIPK3. Indeed, DNMT1 can methylate the RIPK3 promotor, thereby silencing RIPK3 expression and increasing necroptosis resistance of otherwise necroptosis competent cells such as mouse embryonic fibroblasts and L929 cells as well as human HT29 cells [25]. In reverse, RIPK3 promoter demethylation is required and sufficient to restore RIPK3 expression in various human cancer cell models [22]. However, melanoma cells so far were not studied in this context, neither were treatment conditions used that would unequivocally confirm that restoring RIPK3 expression is sufficient to enhance canonical necroptosis signal transduction. We demonstrate that increasing intrinsic RIPK3 expression by demethylating agent 5-AD is insufficient to establish necroptosis competency in melanoma, highlighting that additional regulatory processes must play a role in suppressing necroptosis competency in this setting.
One obvious reason might be that in addition to insufficient RIPK3 also reduced MLKL expression limits necroptosis in melanoma. Indeed, MLKL expression correlated with necroptosis susceptibility in a large cell line-based screen [21], and we here found that MLKL is heterogeneously expressed in melanoma cell lines. However, we showed that melanoma cells expressing MLKL failed to undergo necroptosis, even in presence of elevated RIPK3 amounts. MLKL expression increased after 5-AD pre-treatment in Malme-3M and MEL-JUSO cells, but these cells also remained necroptosis resistant. It is noteworthy, though, that the increase in MLKL after 5-AD treatment indicates that also MLKL expression could be silenced by DNA methylation. To our knowledge, this has not yet been studied, but would align with the more general hypothesis that disease progression in cancer might select for increased necroptosis resistance [21].
We found that signal transduction towards necroptosis never proceeds past RIPK1, indicating that additional regulatory processes must exist. We identified TAK1 as a critical regulator of cell death susceptibility in response to activating TNF receptors and inhibiting cellular IAPs. TAK1 is well known to modulate both apoptosis and necroptosis susceptibility [14,15,16], but the consequences of interfering with TAK1 activity appear to be complex and possibly context dependent. In necroptosis competent fibroblasts, the inhibition of TAK1 suppresses pro-survival NFκB signaling in response to TNFα, augments RIPK1 phosphorylation, and thereby strengthens both apoptosis and necroptosis signaling branches [14]. Furthermore, the extent by which TAK1 directly phosphorylates RIPK1 can regulate apoptosis vs. necroptosis signaling and cell death. If NFκB signaling is suppressed, transient RIPK1 Ser321 phosphorylation supports RIPK1-independent apoptosis, whereas sustained multi-site phosphorylation of RIPK1 supports necroptosis in L929 cells and MEFs [15]. Irrespective of this and regardless of whether TAK1 is inhibited or not, it remains to be studied why melanoma cells exclusively direct signaling towards apoptosis rather than necroptosis, even though all key necroptosis effectors are in place and treatment scenarios otherwise sufficient to induce necroptosis were chosen. Of note, we observed in TRAIL treatment scenarios that cell death remained completely independent of RIPK1 activity, even though TAK1 can regulate TRAIL responsiveness [39, 40]. During TRAIL-induced signaling, a cell death-suppressing role of TAK1 might be less apparent since the apoptosis branch, in contrast to TNFα-induced cell death, relies less on forming an intracellular complex II [41, 42]. Since TAK1 also regulates interleukin and toll-like receptor pathway signaling [43], interfering with the basal activities of these pathways through targeting TAK1 might have contributed to cell death sensitization in our studies.
We found that high TAK1 expression correlates with a higher risk of disease recurrence in patients diagnosed with primary melanoma. Since TNFα is found within melanoma microenvironments [10, 37], TAK1 might contribute to suppressing cell death in situ and thereby promote disease progression. Of note, TNFα is given in combination with DNA-alkylating agents to exert anti-tumor activity during the treatment of loco-regionally metastatic melanoma by isolated limb perfusion [44]. This treatment regimen is not tolerated systemically but clearly of benefit to many patients. Suppressing TAK1 activity with specific inhibitors [28] could, however, provide opportunities to improve these therapeutic strategies in the future.
References
Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39.
Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14:463–82.
Vanden Berghe T, Kaiser WJ, Bertrand MJ, Vandenabeele P. Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Mol Cell Oncol. 2015;2:e975093.
Lafont E, Hartwig T, Walczak H. Paving TRAIL’s path with ubiquitin. Trends Biochem Sci. 2018;43:44–60.
Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–41.
Galluzzi L, Kepp O, Chan FK, Kroemer G. Necroptosis: mechanisms and relevance to disease. Annu Rev Pathol. 2017;12:103–30.
Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54:133–46.
Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278:51613–21.
Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007;19:2056–67.
Smith MP, Sanchez-Laorden B, O’Brien K, Brunton H, Ferguson J, Young H, et al. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFalpha. Cancer Discov. 2014;4:1214–29.
Bertrand F, Montfort A, Marcheteau E, Imbert C, Gilhodes J, Filleron T, et al. TNFalpha blockade overcomes resistance to anti-PD-1 in experimental melanoma. Nat Commun. 2017;8:2256.
Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B, et al. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol. 2009;187:1037–54.
Fulda S. Molecular pathways: targeting inhibitor of apoptosis proteins in cancer—from molecular mechanism to therapeutic application. Clin Cancer Res. 2014;20:289–95.
Guo X, Yin H, Chen Y, Li L, Li J, Liu Q. TAK1 regulates caspase 8 activation and necroptotic signaling via multiple cell death checkpoints. Cell death Dis. 2016;7:e2381.
Geng J, Ito Y, Shi L, Amin P, Chu J, Ouchida AT, et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun. 2017;8:359.
Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ. 2014;21:1667–76.
Geserick P, Wang J, Schilling R, Horn S, Harris PA, Bertin J, et al. Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell death Dis. 2015;6:e1884.
Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–6.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE. 2012;7:e51862.
Najafov A, Zervantonakis IK, Mookhtiar AK, Greninger P, March RJ, Egan RK, et al. BRAF and AXL oncogenes drive RIPK3 expression loss in cancer. PLoS Biol. 2018;16:e2005756.
Koo GB, Morgan MJ, Lee DG, Kim WJ, Yoon JH, Koo JS, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25:707–25.
Feoktistova M, Wallberg F, Tenev T, Geserick P, Leverkus M, Meier P. Techniques to distinguish apoptosis from necroptosis. Cold Spring Harb Protoc. 2016;2016:pdbtop070375.
Hutt M, Marquardt L, Seifert O, Siegemund M, Muller I, Kulms D, et al. Superior properties of Fc-comprising scTRAIL fusion proteins. Mol Cancer Ther. 2017;16:2792–802.
Yang Z, Jiang B, Wang Y, Ni H, Zhang J, Xia J, et al. 2-HG inhibits necroptosis by stimulating DNMT1-dependent hypermethylation of the RIP3 promoter. Cell Rep. 2017;19:1846–57.
Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2’-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–24.
Wu J, Powell F, Larsen NA, Lai Z, Byth KF, Read J, et al. Mechanism and in vitro pharmacology of TAK1 inhibition by (5Z)-7-oxozeaenol. ACS Chem Biol. 2013;8:643–50.
Kilty I, Jones LH. TAK1 selective inhibition: state of the art and future opportunities. Future Med Chem. 2015;7:23–33.
Schadendorf D, Moller A, Algermissen B, Worm M, Sticherling M, Czarnetzki BM. IL-8 produced by human malignant melanoma cells in vitro is an essential autocrine growth factor. J Immunol. 1993;151:2667–75.
Bar-Eli M. Role of interleukin-8 in tumor growth and metastasis of human melanoma. Pathobiology. 1999;67:12–18.
Richmond A, Thomas HG. Melanoma growth stimulatory activity: isolation from human melanoma tumors and characterization of tissue distribution. J Cell Biochem. 1988;36:185–98.
Dhawan P, Richmond A. Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol. 2002;72:9–18.
Payne AS, Cornelius LA. The role of chemokines in melanoma tumor growth and metastasis. J Invest Dermatol. 2002;118:915–22.
Stove C, Vanrobaeys F, Devreese B, Van Beeumen J, Mareel M, Bracke M. Melanoma cells secrete follistatin, an antagonist of activin-mediated growth inhibition. Oncogene. 2004;23:5330–9.
Becker JC, Dummer R, Hartmann AA, Burg G, Schmidt RE. Shedding of ICAM-1 from human melanoma cell lines induced by IFN-gamma and tumor necrosis factor-alpha. Functional consequences on cell-mediated cytotoxicity. J Immunol. 1991;147:4398–401.
Dondelinger Y, Aguileta MA, Goossens V, Dubuisson C, Grootjans S, Dejardin E, et al. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 2013;20:1381–92.
Bergenwald C, Westermark G, Sander B. Variable expression of tumor necrosis factor alpha in human malignant melanoma localized by in situ hybridization for mRNA. Cancer Immunol Immunother. 1997;44:335–40.
Santoro R, Carbone C, Piro G, Chiao PJ, Melisi D. TAK-ing aim at chemoresistance: The emerging role of MAP3K7 as a target for cancer therapy. Drug Resist Updat. 2017;33-35:36–42.
Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–85.
Morioka S, Omori E, Kajino T, Kajino-Sakamoto R, Matsumoto K, Ninomiya-Tsuji J. TAK1 kinase determines TRAIL sensitivity by modulating reactive oxygen species and cIAP. Oncogene. 2009;28:2257–65.
Lafont E, Kantari-Mimoun C, Draber P, De Miguel D, Hartwig T, Reichert M, et al. The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J. 2017;36:1147–66.
Jin Z, El-Deiry WS. Distinct signaling pathways in TRAIL-versus tumor necrosis factor-induced apoptosis. Mol Cell Biol. 2006;26:8136–48.
Sakurai H. Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharmacol Sci. 2012;33:522–30.
van Horssen R, Ten Hagen TL, Eggermont AM. TNF-alpha in cancer treatment: molecular insights, antitumor effects, and clinical utility. Oncologist. 2006;11:397–408.
Acknowledgements
We mourn the passing of our thoughtful and enthusiastic co-author, colleague and friend Martin Leverkus. LGP and MR kindly acknowledge support by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement #642295 (MEL-PLEX). MR and DPD also receive support from the German Research Foundation (FOR2036 (MO 3226/1-1), DI 2440/3-1). MR receives further support from the European Union’s Horizon 2020 research and innovation programme (grant agreements #766069 (GLIO-TRAIN), #675448 (TRAIN-ERS)) and the Health Research Board Ireland (HRA POR 2013 245, HRA POR 2015 1091).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Supplementary information
Rights and permissions
About this article
Cite this article
Podder, B., Guttà, C., Rožanc, J. et al. TAK1 suppresses RIPK1-dependent cell death and is associated with disease progression in melanoma. Cell Death Differ 26, 2520–2534 (2019). https://doi.org/10.1038/s41418-019-0315-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0315-8
This article is cited by
-
Comprehensive prediction of immune microenvironment and hot and cold tumor differentiation in cutaneous melanoma based on necroptosis-related lncRNA
Scientific Reports (2023)
-
Housing conditions affect enterocyte death mode and turnover rate in mouse small intestine
Scientific Reports (2023)
-
TAK1 Reduces Surgery-induced Overactivation of RIPK1 to Relieve Neuroinflammation and Cognitive Dysfunction in Aged Rats
Neurochemical Research (2023)
-
MLKL deficiency in BrafV600EPten−/− melanoma model results in a modest delay of nevi development and reduced lymph node dissemination in male mice
Cell Death & Disease (2022)
-
ABIN-1 is a key regulator in RIPK1-dependent apoptosis (RDA) and necroptosis, and ABIN-1 deficiency potentiates necroptosis-based cancer therapy in colorectal cancer
Cell Death & Disease (2021)