Abstract
Premenstrual Dysphoric Disorder (PMDD) is characterized by debilitating mood symptoms in the luteal phase of the menstrual cycle. Prior studies of affected women have implicated a differential response to ovarian steroids. However, the molecular basis of these patients’ differential response to hormone remains poorly understood. We performed transcriptomic analyses of lymphoblastoid cell lines (LCLs) derived from women with PMDD and asymptomatic controls cultured under untreated (steroid-free), estradiol-treated (E2), and progesterone-treated (P4) conditions. Weighted gene correlation network analysis (WGCNA) of transcriptomes identified four gene modules with significant diagnosis x hormone interactions, including one enriched for neuronal functions. Next, in a gene-level analysis comparing transcriptional response to hormone across diagnoses, a generalized linear model identified 1522 genes differentially responsive to E2 (E2-DRGs). Among the top 10 E2-DRGs was a physically interacting network (NUCB1, DST, GCC2, GOLGB1) involved in endoplasmic reticulum (ER)-Golgi function. qRT-PCR validation reproduced a diagnosis x E2 interaction (F(1,24)=7.01, p = 0.014) for NUCB1, a regulator of cellular Ca2+ and ER stress. Finally, we used a thapsigargin (Tg) challenge assay to test whether E2 induces differences in Ca2+ homeostasis and ER stress response in PMDD. PMDD LCLs had a 1.36-fold decrease in Tg-induced XBP1 splicing response compared to controls, and a 1.62-fold decreased response (p = 0.005), with a diagnosis x treatment interaction (F(3,33)=3.51, p = 0.026) in the E2-exposed condition. Altered hormone-dependent in cellular Ca2+ dynamics and ER stress may contribute to the pathophysiology of PMDD.
Similar content being viewed by others
Introduction
Premenstrual Dysphoric Disorder (PMDD) affects 3–8% of reproductive-aged women and involves debilitating mood symptoms confined to the luteal phase of the menstrual cycle [1, 2]. Symptoms occur after ovulation and remit shortly after menses, suggesting a role for luteal hormones. While peripheral levels of estradiol, progesterone, and other reproductive steroids are normal in PMDD [3, 4], clinical hormone manipulations suggest PMDD is a disease induced by physiologic fluctuations in reproductive hormones. Women with PMDD experience remission of mood symptoms after pharmacologic suppression of the hypothalamic-pituitary-gonadal (HPG) axis with GnRH agonists and recurrence when estradiol (E2) or progesterone (P4) are blindly added back—a response not seen in healthy controls [5]. Neuroimaging studies have demonstrated both persistent and hormone-induced differences in neurocircuitry and cognition in PMDD, including emotional processing, corticolimbic control, and working memory [6, 7].
Recent evidence suggests these physiologic characteristics of PMDD may be underpinned by genetic and cellular factors. Using lymphoblastoid cell lines (LCLs), the ESC/E(Z) complex, a set of 13 genes involved in epigenetic silencing via H3K27 methylation, was found to be differentially expressed in PMDD via transcriptomic analysis in the baseline, hormone-free condition [8]. While ESC/E(Z) expression differences could serve as a potential substrate for an abnormal response to hormone, the molecular and functional correlates of a differential hormone response (including the transcriptional consequences of altered ESC/E(Z) complex expression) have not been fully characterized.
In this study, we first explore the transcriptomic basis of differential steroid response in PMDD using a module-level analysis to identify differentially regulated gene modules and a gene-level analysis to identify differentially regulated genes. Informed by these results, we designed an in-vitro assay to test whether there was a hormone-dependent functional difference in Ca2+ homeostasis and ER stress response between PMDD and control LCLs.
Methods
Participants
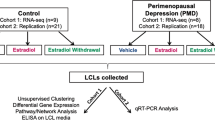
Women with PMDD and asymptomatic controls were recruited as previously described [8]. Briefly, participants were 18-48 years old, had regular menses (21–35 days), and were not medically ill, taking medications, or pregnant. DSM-5 criteria were prospectively confirmed through daily symptom ratings over three menstrual cycles [9]. Women underwent clinical interview and self-report questionnaire to confirm diagnostic criteria. Additionally, each woman had symptom ratings (irritability, depression, anxiety, mood lability) ≥30% increased (relative to the range of the symptom scale) in the week before menses, compared to the week after. Women with significant symptoms outside the luteal phase were excluded.
In controls, absence of symptoms was confirmed using daily ratings over 2 months. Women contributing samples for RNAseq underwent a trial of leuprolide and hormone add-back [8]. Women in the PMDD RNAseq group experienced symptom remission with leuprolide and recurrence with E2 or P4 add-back using defined criteria [5]. The control group remained euthymic throughout the entire protocol. An independent cohort of women with PMDD and controls who otherwise met study inclusion criteria but did not undergo leuprolide/add-back trials contributed samples for experimental validation. Participant demographics summarized in Supplemental Table S1. This study was approved by the NIH IRB. All participants provided written informed consent according to NIH intramural guidelines.
Cell culturing, hormone treatments, and RNA sequencing
LCLs were generated via Epstein-Barr Virus transformation of mononuclear cells from peripheral blood [10]. LCLs at 2 × 105 cells/mL in phenol-free RPMI (Gibco, cat. 11835-055) with 15% KOSR (Gibco, cat. 10828-028), 2% glutamine (200 mM), and 1% gentamicin (5 mg/ml) were cultured for 3–5 days at 37 °C with 5% CO2 before treatment with estradiol (100 nM, Sigma-Aldrich, cat. E2758-5G) or progesterone (100 nM, Sigma-Aldrich, cat. P8783-25G) for 24 h, or no treatment. Full details are given in Dubey et al. [8]. Cells were harvested for RNAseq, described in Supplemental Methods. Workflow of data generation and analysis is summarized in Supplemental Fig. S1.
Module-level analysis: WGCNA, GSEA, and Enrichr
Weighted Gene Correlation Network Analysis (WGCNA) was conducted using R v.1.66 [11]. Modules were generated from CPM values of 46 samples analyzed by RNAseq. To identify modules differentially responsive to hormone in PMDD, two-way ANOVA was performed on module eigengenes, and modules showing a diagnosis x hormone interaction (nominal P < 0.05) were considered differentially responsive. Functional analysis of modules was performed using Gene Set Enrichment Analysis (GSEA) [12]. In a secondary analysis, core subsets underlying top GSEA enrichment signals were defined using Leading Edge Analysis (LEA), and a consensus set was used for pathway analysis under Enrichr tools [13]. Full details are given in Supplemental Methods.
Gene-level analysis: EdgeR, GeneMANIA, and qRT-PCR
Generalized linear modeling of RNAseq data within EdgeR identified genes differentially regulated by either E2 (E2-DRGs) or P4 (P4-DRGs) [14]. Each set of paired RNAseq samples (untreated and E2-treated, and untreated and P4-treated) was used to fit a separate model of hormone response. Paired samples with Trimmed Mean of M-Values (TMM)-normalized counts and tagwise dispersion estimation were fitted to a quasi-likelihood negative binomial generalized log-linear model. Factors of subject (cell line) and treatment (untreated vs. hormone-treated) were nested under diagnosis (control vs. PMDD). Genewise F-tests were performed with each model to identify genes showing significant diagnosis x hormone interactions (nominal P < 0.05), yielding a list of E2-DRGs and P4-DRGs.
Network visualization was performed in GeneMANIA, a web-based tool that accepts an unranked gene list and generates a weighted interaction network based on published databases [15]. Select genes were validated in a biologically independent cohort of cases and controls via TaqMan qRT-PCR (Applied Biosystems), described in Supplemental Methods.
Thapsigargin challenge assays
A thapsigargin (Tg) challenge assay compared the response of PMDD and control LCLs to Tg, a sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) channel inhibitor, under untreated and E2-treated conditions. LCLs were seeded at 2 × 105 cells/mL in phenol red-free RPMI, divided into 5 ml cultures in 6-well plates, and cultured for 3–5 days. For E2 pre-treatments, E2 in DMSO was added for a final concentration of 100 nM and cultured for 20 h. An equivalent volume of DMSO served as vehicle control. For Tg treatments, Tg (DMSO) and CaCl2 (aq) were added for a final concentration of 0.2 uM and 0.8 uM, respectively, and cultured for an additional 4 h. RNA extraction and cDNA synthesis were performed as described above. Spliced-to-unspliced XBP1 ratios (s/u XBP1), a marker of ER stress response activation, were quantified via qRT-PCR, described in Supplemental Methods.
Statistical analysis
Statistical analyses were performed in GraphPad Prism v8 (La Jolla, CA) and R v3.4.1 (Vienna, Austria). Student’s t test and Fisher’s Exact tests were conducted for demographic variables. qRT-PCR analyses were performed with repeated measures ANOVA with Sidak’s multiple comparisons.
Results
WGCNA identifies modules associated with abnormal hormone response
Of 26,362 genes represented in 46 input libraries, 22,242 (84%) were sufficiently expressed for WGCNA, and 21,746 (98%) clustered into 65 modules (Fig. 1a). Four modules (arbitrarily designated Turquoise, Pink, Brown, and Magenta) had eigengenes with significant diagnosis x hormone interactions (Fig. 1b, Supplemental Table S2).
a Cluster dendrogram of gene modules. In WGCNA analysis of 46 input transcriptomes, 21,746 unique gene features (“genes”) were clustered into 65 co-expression modules using gene–gene correlation patterns across all input transcriptomes. b Module eigengenes showing significant diagnosis x hormone interactions. Eigengenes (defined as the first principal component of a given module) of four modules (Brown, Pink, Turquoise, and Magenta) showed differential regulation by hormone (ANOVA interaction P < 0.05). Closed circles denote controls. Open circles denote PMDD. Note: An extreme outlier in the E2-treated control group was excluded in the eigengene plot of the Magenta module for scaling purposes. Hormone x diagnosis interaction was significant both before and after exclusion of this outlier. P-values are not adjusted for multiple comparisons. c Module membership of ESC/E(Z) complex genes. Nearly half (6/13) of ESC/E(Z) complex genes were assigned to the Turquoise module. Remaining ESC/E(Z) genes were clustered into the Blue, AntiqueWhite4 (aw4), DarkOliveGreen (dog), Cyan (cy), Green (g), and SkyBlue2 (sb2) modules. d ESC/E(Z) complex genes in the Turquoise module. Genes plotted according to module membership and strength of association with diagnosis. ESC/E(Z) complex genes (SUZ12, EED, AEBP2, HDAC2, MTF2, SIRT2) are more likely to have high module membership within the Turquoise module. e Word cloud representations of GSEA results. Top 20 GSEA results were used to generate word clouds for each differentially regulated module. Word size is proportional to frequency of word among GO terms identified by GSEA results. Modules were enriched for terms relating to MAP Kinase and GTPase signaling (Brown, top left), chromatin remodeling and proteasome function (Turquoise, top right), mitochondrial function (Pink, bottom left), and neuronal and synaptic function (Magenta, bottom right). Full, quantitative GSEA results are tabulated in Supplemental Table S3. Word clouds generated via worditout.com.
All 13 ESC/E(Z) complex genes clustered into modules with significant effects of diagnosis, hormone, or their interaction (Fig. 1c). Nearly half (SIRT1, MTF2, AEBP2, HDAC2, SUZ12, EED) were high-connectivity members of the Turquoise module. SUZ12 had the fourth highest connectivity by module membership among 2492 module genes (Fig. 1d).
In functional GSEA analysis of significant modules, Turquoise was enriched for terms relating to chromatin remodeling, proteasome function, and transcriptional processing. Pink was enriched for mitochondrial function. Brown contained many terms for MAP Kinase and GTPase signaling. Magenta was enriched for neuronal and synaptic function (Fig. 1e). Complete GSEA results in Supplemental Table S3.
Top E2-DRGs comprise a physically interacting network involved in ER–Golgi function and Ca2+ homeostasis
Linear modeling of RNAseq data in EdgeR identified 1522 genes differentially responsive to E2 (E2-DRGs) and 480 differentially responsive to P4 (P4-DRGs). Full EdgeR results in Supplemental Tables S4, S5.
We focused on the E2-treated condition because: (1) E2-treatment appeared to drive the diagnosis x hormone interaction seen in modules of interest (Fig. 1b); (2) There were no P4-DRGs with FDR < 0.25 (Supplemental Table S5); (3) Hierarchical clustering of transcriptomes showed stronger clustering among E2-treated samples compared to P4-treated samples, and greater segregation between cases and controls within the E2-treated cluster (Supplemental Fig. S2a); and (4) Modules of interest tended to be enriched for E2-DRGs rather than P4-DRGs (Supplemental Fig. S2b, Supplemental Table S2).
Table 1 lists the top 10 E2-DRGs. Several of these genes are directly related to cellular Ca2+ handling and the ER–Golgi compartment (NUCB1, DST, GCC2, and GOLGB1), and were included as inputs for GeneMANIA network visualization. The resulting output revealed a network of Golgi–ER-associated genes, with physical interactions contributing 67.6% to the weighted network (Fig. 2a).
a GeneMANIA visualization of select E2-DRGs. Key genes relating to ER–Golgi function and intracellular calcium homeostasis (NUCB1, DST, GCC2, GOLGB1) were selected from among the top 10 E2-DRGs and included as inputs for GeneMANIA network visualization (genemania.org, accessed January 2019). The resulting output revealed a structural network of Golgi–ER-associated genes, with physical interactions comprising 67.6% of the weighted network. b qRT-PCR validation of select E2-DRGs. NUCB1, DHCR7, PPP2R5D, and GOLGB1 were selected for qRT-PCR validation in an independent replication cohort of 13 cases and 13 controls. NUCB1 qRT-PCR results revealed a significant main effect of diagnosis (F(1,24)=4.61, p = 0.042) and interaction of diagnosis and hormone (F(1,24)=7.01, p = 0.014). In post-hoc testing, E2-treated PMDD samples had significantly reduced NUCB1 expression compared to control samples (p = 0.003). qRT-PCR results for DHCR7 did not reveal significant effects of diagnosis, hormone, or their interaction, but in multiple comparisons testing, DHCR7 expression was significantly reduced in E2-treated PMDD samples compared to E2-treated control samples (p = 0.040). No significant differences for PPP2R5D or GOLGB1 were found in qRT-PCR validation.
Select genes were chosen for qRT-PCR validation in a biologically independent cohort of 13 cases and 13 controls. Repeated measures ANOVA on NUCB1 qRT-PCR results revealed a main effect of diagnosis (F(1,24)=4.61, p = 0.042) and diagnosis x hormone interaction (F(1,24)=7.01, p = 0.014) (Fig. 2b).
E2 induces differences in Ca2+ homeostasis and ER stress response in PMDD LCLs
Thapsigargin (Tg), a potent SERCA inhibitor, rapidly increases cytosolic Ca2+ levels and triggers the ER stress response [16]. We tested the effects of Tg and E2, alone and in combination, and found a significant main effect of treatment (F(3,33)=11.2, p < 0.001) and a significant diagnosis x treatment interaction (F(3,33)=3.51, p = 0.026). The main effect of diagnosis was itself not significant (p = 0.064) (Fig. 3a).
a Experimental design of Tg challenge assays. PMDD and control LCLs were cultured in phenol red-free RPMI for 5–6 days and treated in four arms: (1) no treatment (vehicle), (2) E2 (100 nM) for 24 h, (3) Tg (0.2 uM) and CaCl2 (0.8 mM) for 4 h, and (4) E2 (100 nM) for 24 h with Tg (0.2 uM) and CaCl2 (0.8 mM) for the last 4 h. Cells were harvested and assayed for the spliced-to-unspliced ratio of XBP1 by qRT-PCR. b XBP1 S/US ratios in Tg challenge assays (by diagnosis group). Control LCLs showed an increase in XBP1 S/US ratio after Tg treatment. Pre-treatment with E2 potentiated this response. Both of these effects were blunted or absent in PMDD LCLs. A significant diagnosis x treatment interaction (ANOVA-RM F(3,33) = 3.508, p = 0.026) in XBP1 splicing response to Tg was observed. Post-hoc analysis revealed a significant difference between control and PMDD LCLs only when cells were treated with the combination of E2 + Tg (p = 0.005). E2 had a significant modulatory effect on Tg response in control cells (p = 0.007) but not in PMDD cells (p = 0.693) (see Supplemental Table S6). c S/US XBP1 ratios in Tg challenge assays (by individual cell line). Examining S/US XBP1 ratios of individual cell lines across treatment conditions shows qualitatively distinct trajectories between control and PMDD groups, most notably, the loss of E2-mediated potentiation of XBP1 splicing in PMDD LCLs. However, significant within-group heterogeneity among cell lines is also appreciated.
In post-hoc analysis, control LCLs had a 1.7-fold increase in XBP1 splicing with Tg alone (p = 0.009) and a 2.2-fold increase with Tg preceded by E2 (p < 0.0001). PMDD LCLs showed a blunted, non-significant increase in XBP1 splicing in both conditions. Furthermore, while E2 exposure modulates Tg-induced XBP1 splicing in Control LCLs (p = 0.007), addition of E2 did not alter response to Tg in PMDD LCLs (p = 0.693). Comparing PMDD and control LCLs within-treatment, XBP1 splicing in PMDD was 1.36-fold decreased compared to controls in the Tg-only condition (p = 0.25). This difference was accentuated (1.62-fold decrease, p = 0.005) when Tg was preceded by E2. E2 alone did not affect XBP1 splicing regardless of diagnosis (Fig. 3b, c). Full ANOVA-RM results in Supplemental Table S6.
Secondary analysis of a neuronally relevant module implicates Ca2+ dynamics and the ESC/E(Z) complex
We revisited our module-level analysis to see if Ca2+-related pathways were implicated by Magenta module genes. To explore potential pathways represented by Magenta module hub genes, Leading Edge Analysis (LEA) of top 20 enriched Magenta GO terms with FDR < 0.25 was used to identify a core subset of 88 genes (Supplemental Table S7). Enrichr analysis found that these genes were enriched for GO terms relating to Ca2+ dynamics, including Ca2+-dependent synaptic vesicle handling and Ca2+ ion transport. Surprisingly, these genes were also enriched for targets of ESC/E(Z) complex genes SUZ12 and EZH2, estrogen receptor ESR1, and RE1-Silencing Transcription Factor (REST), which suppresses expression of neural genes in non-neuronal cell lines [17], and genes associated with the H3K27me3 histone marker. The top enriched KEGG pathways were neuroactive ligand–receptor interaction, the cAMP, Rap1, and Ras pathways, and glutamatergic synapse (Supplemental Fig. S3). Full Enrichr results are given in Supplemental Table S8.
Discussion
PMDD is a disorder of atypical response to ovarian steroids [5]. While we recently reported differences in ESC/E(Z) expression in hormone-free conditions, the molecular and functional correlates of differential hormone response in PMDD remain poorly defined. Comparing LCLs from PMDD cases and controls, we identified four gene modules differentially responsive to hormone, including one enriched for neuronal functions. Analysis of E2-DRGs suggested a role for altered Ca2+ handing and the ER–Golgi system. A functional assay showed both the ER stress response and its normal modulation by E2 were blunted in PMDD. Thus, starting from a global transcriptomic analysis, we report the first indications of a hormone-dependent functional difference in cells of women with PMDD.
Each of four modules implicated via WGCNA represents a plausible pathway in PMDD. Two modules (Blue and Pink) were enriched for MAPK and GTPase signaling and mitochondrial function, respectively. These functions have been implicated in mood disorders including as mediators of sex differences [18,19,20,21,22] and estrogen response [23,24,25]. The Turquoise module is associated with ESC/E(Z), an epigenetic modifier gene complex recently implicated by our group in PMDD. Isolation of a neuronally relevant module (Magenta) is notable because LCLs are non-neuronal cells. While other LCL-based studies of psychiatric disease identified individual genes of potential neuronal relevance [26,27,28,29], with others leveraging WGCNA to implicate non-specific cellular or immunologic modules [30,31,32], this is the first study using LCLs to implicate a neuronally relevant module in psychiatric disease. In a preliminary analysis, Magenta hub genes were enriched for targets of REST, a repressor of neuronal genes in non-neuronal cells [17], suggesting that despite mechanisms limiting neuronal gene expression such as REST-mediated transcriptional repression, potentially meaningful differences in aggregate expression could still be detected, perhaps due to careful phenotyping and appropriate clinical stratification.
Our gene-wise analysis implicated the ER–Golgi system and cellular Ca2+ dynamics. NUCB1, an independently validated E2-DRG, encodes a Golgi protein regulating endomembrane Ca2+ uptake and ER stress response [33, 34]. It is also implicated in spatiotemporal Ca2+ regulation during neurotransmission [35], with an emerging role in mood and behavior. NUCB1 is modulated by sex hormones and affects behavior in animal models [36]. Homolog NUCB2 is a sex-specific correlate of depression, anxiety, and suicide in humans [37,38,39,40]. Other top E2-DRGs include GOLGB1 (encodes a cross-bridging protein of Golgi cisternae), GCC2 (encodes a molecular tether between transport vesicles and the Golgi), and DST (encodes a neuronal isoform critical in ER function) [41,42,43]. DST mutations cause hereditary sensory neuropathy, possibly via Ca2+ dyshomeostasis, pathologic ER stress response induction, and subsequent sensory neuron apoptosis [44]. Together, the differential expression of these genes suggests Ca2+ homeostasis and its effects on the Golgi–ER system and ER stress could be disrupted in PMDD.
The ER stress response, also known as the unfolded protein response, is a highly conserved response to Ca2+ perturbation and protein misfolding: induction of protein-folding chaperones, ribosome halting, upregulated proteolysis, and increased cytoplasmic-endomembrane Ca2+ transport [45, 46]. In the LCL model, E2 (rather than P4) was the primary driver of differential hormone response, and was thus used in our functional assays. While P4 is clinically important [47, 48], the primacy of E2 in our RNAseq experiments likely reflects the absence of nuclear P4 receptors in lymphoid cells [8, 49]. On the other hand, nuclear estrogen receptors ESR1 and ESR2 are abundantly expressed and mediate estrogen-dependent changes in gene expression [50]. These are distinct from membrane E2 and P4 receptors, which are both expressed in lymphoid cells but are thought to mediate the immunomodulatory effects of E2 and P4 via rapid intracellular signal transduction events rather than transcriptional regulation [51, 52]. Furthermore, while both membrane steroid receptors and nuclear steroid receptors (in their membrane-localized state) likely orchestrate E2- and P4-mediated synaptic remodeling via non-genomic signal transduction events, particularly via G-protein coupled receptors, these effects are less likely to be detected by an RNAseq-based methodology [53,54,55,56].
Although control LCLs initiate a Tg-induced ER stress response that is potentiated by E2, both of these effects are diminished or absent in PMDD LCLs. This result parallels similar findings in LCLs from patients with bipolar disorder (BD). So et al. found attenuated XBP1 induction in BD LCLs compared to controls [57]. Similar findings were later independently reported by Hayashi et al. and Pfaffenseller et al. [58, 59]. Interestingly, BD shares significant phenotypic overlap with PMDD, and PMDD patients are frequently misdiagnosed with BD [60].
While gene-level analysis identified select E2-DRGs related to Ca2+ and ER stress, the connection between module-level findings and our observed impairments in ER stress response was less clear. Due to the potential relevance of Ca2+ dynamics to neuronal biology, we wondered if a secondary analysis of the Magenta module would implicate Ca2+-relevant pathways. As GSEA results suggested broad functional terms (“Synapse”, “Neuron”, “Axon”) without specifically implicating subcellular pathways, we focused on a union set of 88 leading edge (LEA) genes compiled from top GSEA enrichment signals. LEA subsets prioritize high-rank (i.e., high-connectivity) genes, selecting for the most functionally relevant hub genes. In addition to identifying enriched GO terms relating to Ca2+ handling, Enrichr analysis of this subset also implicated targets of ESC/E(Z) genes SUZ12 and EZH2 and estrogen receptor ESR1, suggesting a potential relationship between the ESC/E(Z) complex, estrogen response, and transcriptional control of Magenta module genes. This relationship is reinforced by a concomitant enrichment of histone marker H3K27me3-associated genes; H3K27 is a specific target of ESC/E(Z) [61], and the only known methyltransferase capable of methylation at H3K27me3 is EZH2, a core ESC/E(Z) effector [62]. As this implies a regulatory relationship between Magenta and Turquoise modules, both differentially responsive modules in PMDD, we hypothesize that in PMDD, a set of genes involved in neurotransmission, synaptic function, and Ca2+ dynamics (represented by the Magenta module) may be abnormally regulated by H3K27 epigenetic remodeling (represented by the Turquoise module and the ESC/E(Z) complex). These putative relationships require further study.
There is some genetic evidence for ER stress genes in neuropsychiatric disease. Among canonical ER stress response mediators XBP1, GRP94 (HSP90B1), GRP78 (HSPA5), CHOP (DDIT3), PERK (EIF2AK3), ATF6, and IRE1 (ERN1), Genome-wide association (GWA) studies identified a PERK variant (rs7571971) associated with progressive supranuclear palsy [63], and an ATF6 variant (rs10918270) associated with Parkinson’s Disease [64]. The plausible implication of ER stress pathways in PMDD by RNAseq suggests that transcriptomics may uncover genetic etiologies not revealed by GWAS, which has only partially explained the liability to affective illnesses.
E2 has been studied as a modulator of ER stress. In breast cancer, E2 has a protective, activating effect on ER stress response. Under mitogenic E2 signaling, cells activate an “anticipatory” ER stress response alongside upregulated protein synthesis and mitosis [65, 66]. In non-dividing cells such as neurons, E2-mediated priming of ER stress pathways might protect against non-mitotic sources of ER stress and Ca2+ perturbation—namely, excitatory synaptic input. This is consistent with prior work demonstrating protective effects of estrogen in glutamate excitotoxicity via Ca2+ dynamics stabilization [67]. Blunting of this anticipatory homeostatic pathway may increase neuronal excitability and partially explain the observation that women with PMDD demonstrate a relative resistance to the expected inhibitory effects of GABA-A receptor modulators such as allopregnanolone, thought to be a key pathophysiologic mechanism in PMDD [68]. Cellular Ca2+ dynamics are also critical in microglial activation, a neuroimmune process perhaps better approximated by LCLs. Dysregulated microglial activation and other neuroinflammatory processes have been implicated in depressive disorders [69, 70]. Finally, a critical end-organelle of ER stress signaling are mitochondria, which play important roles in Ca2+ sequestration, ROS metabolism, inflammatory signaling, and apoptosis [71, 72]. Mitochondria were not only implicated in our own WGCNA analysis, but have emerged as mediators of estrogen’s neuroactive effects in psychiatric and neurodegenerative disease, particularly through their role in regulating intracellular Ca2+ [24, 71, 73, 74].
Our findings also have therapeutic implications. The ER stress pathway is altered by many mood stabilizing drugs [75], and there is ample clinical precedent for targeting cellular Ca2+ in psychiatric disease. Although now supplanted by antidepressants and mood stabilizers, Ca2+ channel blockers such as verapamil were once commonly and efficaciously used in the treatment of anxiety, depression, bipolar disorder, and premenstrual symptoms [76, 77]. If confirmed in future studies, hormone-dependent alterations in the ER stress response and Ca2+ homeostasis represent a novel yet conceptually plausible pathway in the pathophysiology of PMDD.
Limitations and future work
We found hormone-dependent differences in Tg-induced ER stress response, but in a modest sample of 13 patient-derived LCLs. Cell lines used in Tg experiments were not completely independent from those used in initial RNAseq experiments. Generalizability of results would be supported by validation in larger, independent cohorts.
Our findings of altered ER stress require mechanistic follow-up. By using XBP1 splicing as the functional output, we did not investigate Ca2+ homeostasis and ER stress as independent processes. Quantifying Ca2+ dynamics through calcium imaging or electrophysiology, and measuring other ER stress markers (e.g., GRP94, GRP78, CHOP) or ER stress intermediaries (PERK, ATF6, IRE1) would enable a better mechanistic understanding of altered Ca2+ dynamics and ER stress in PMDD. The role of NUCB1 in modulating ER stress and Ca2+—a promising lead given its validation as an E2-DRG—is another area for future experimentation.
The role of ESC/E(Z) in mediating this cellular phenotype should also be investigated. A secondary analysis of key Magenta module hub genes suggested a role for ESC/E(Z) in regulating both neuronal function and cellular Ca2+ dynamics. The selection of Leading Edge genes for secondary analysis, while an established method of isolating statistically and functionally important genes within a module, can introduce selection bias. We cannot conclude that this module is the transcriptional basis of altered Ca2+ homeostasis. The purported role of ESC/E(Z) on Ca2+ dynamics and neuronal biology may be better addressed by multi-omic methods (integration of ChIP-seq and RNAseq datasets) and the use of a more neuronally relevant model (e.g., neuronal cell lines from patient-derived stem cells or postmortem brain tissue), especially as LCLs are in many respects not an authentic model of neuropsychiatric function [78,79,80]. For PMDD, a particular gap is the absence of nuclear progesterone receptors in LCLs. Furthermore, attribution of our findings to genetic variants or epigenetic markers would extend the understanding of PMDD as a genetically influenced disease.
Finally, despite significant differences in between-group analysis, appreciable within-group variation existed in NUCB1 expression and ER stress activation in both PMDD and control LCLs. If PMDD is a heterogeneous diagnosis, blunted E2-dependent ER stress activation may be common among both cases and controls, but associated with just one of multiple molecular etiologies resulting in psychiatric vulnerability to luteal phase hormones. Future work to correlate alterations in Ca2+ dynamics and ER stress, with clinical features (including response to therapies targeting these pathways) may allow for stratification of this complex diagnosis for research and clinical management.
References
Halbreich U, Borenstein J, Pearlstein T, Kahn LS. The prevalence, impairment, impact, and burden of premenstrual dysphoric disorder (PMS/PMDD). Psychoneuroendocrinology 2003;28:1–23.
Hofmeister S, Bodden S. Premenstrual syndrome and premenstrual dysphoric disorder. Am Fam Physician 2016;94:236–240.
Schiller CE, Johnson SL, Abate AC, Schmidt PJ, Rubinow DR. Reproductive steroid regulation of mood and behavior. Compr Physiol 2016;6:1135–1160.
Rubinow DR, Schmidt PJ, Roca CA. Hormone measures in reproductive endocrine-related mood disorders: diagnostic issues. Psychopharmacol Bull 1998;34:289–290.
Schmidt PJ, Nieman LK, Danaceau MA, Adams LF, Rubinow DR. Differential behavioral effects of gonadal steroids in women with and in those without premenstrual syndrome. N Engl J Med 1998;338:209–216.
Baller EB, Wei S-M, Kohn PD, Rubinow DR, Alarcón G, Schmidt PJ, et al. Abnormalities of dorsolateral prefrontal function in women with premenstrual dysphoric disorder: a multimodal neuroimaging study. Am J Psychiatry 2013;170:305–314.
Toffoletto S, Lanzenberger R, Gingnell M, Sundström-Poromaa I, Comasco E. Emotional and cognitive functional imaging of estrogen and progesterone effects in the female human brain: a systematic review. Psychoneuroendocrinology 2014;50:28–52.
Dubey N, Hoffman JF, Schuebel K, Yuan Q, Martinez PE, Nieman LK, et al. The ESC/E(Z) complex, an effector of response to ovarian steroids, manifests an intrinsic difference in cells from women with premenstrual dysphoric disorder. Mol Psychiatry 2017;22:1172–1184.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-5®). American Psychiatric Pub; 2013.
Oh H-M, Oh J-M, Choi S-C, Kim S-W, Han W-C, Kim T-H, et al. An efficient method for the rapid establishment of Epstein-Barr virus immortalization of human B lymphocytes. Cell Prolif 2003;36:191–197.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma 2008;9:559.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–97.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl 2010;26:139–140.
Warde-Farley D, Donaldson S, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38. https://pubmed.ncbi.nlm.nih.gov/20576703/. Accessed 16 Sept 2020.
Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 1999;13:1211–1233.
Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet 1998;20:136–142.
Einat H, Yuan P, Gould TD, Li J, Du J, Zhang L, et al. The role of the extracellular signal-regulated kinase signaling pathway in mood modulation. J Neurosci Off. J Soc Neurosci 2003;23:7311–7316.
Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, et al. A negative regulator of MAP kinase causes depressive behavior. Nat Med 2010;16:1328–1332.
Zhang L, Li B Shing, Zhao W, Chang YH, Ma W, Dragan M, et al. Sex-related differences in MAPKs activation in rat astrocytes: effects of estrogen on cell death. Brain Res Mol Brain Res. 2002;103:1–11.
Shimamoto A, Rappeneau V. Sex-dependent mental illnesses and mitochondria. Schizophr Res 2017;187:38–46.
Allen J, Romay-Tallon R, Brymer KJ, Caruncho HJ, Kalynchuk LE. Mitochondria and mood: mitochondrial dysfunction as a key player in the manifestation of depression. Front Neurosci 2018;12:386.
Segars JH, Driggers PH. Estrogen action and cytoplasmic signaling cascades. Part I: membrane-associated signaling complexes. Trends Endocrinol Metab Tem 2002;13:349–354.
Nilsen J, Brinton RD. Mitochondria as therapeutic targets of estrogen action in the central nervous system. Curr Drug Targets CNS Neurol Disord 2004;3:297–313.
Irwin RW, Yao J, Hamilton RT, Cadenas E, Brinton RD, Nilsen J. Progesterone and estrogen regulate oxidative metabolism in brain mitochondria. Endocrinology 2008;149:3167–3175.
Milanesi E, Voinsky I, Hadar A, Srouji A, Maj C, Shekhtman T, et al. RNA sequencing of bipolar disorder lymphoblastoid cell lines implicates the neurotrophic factor HRP-3 in lithium’s clinical efficacy. World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2017;1–13.
Duan J, Göring HHH, Sanders AR, Moy W, Freda J, Drigalenko EI, et al. Transcriptomic signatures of schizophrenia revealed by dopamine perturbation in an ex vivo model. Transl Psychiatry. 2018;8.
Kittel-Schneider S, Hilscher M, Scholz C-J, Weber H, Grünewald L, Schwarz R, et al. Lithium-induced gene expression alterations in two peripheral cell models of bipolar disorder. World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2017;1–14.
Sanders AR, Göring HHH, Duan J, Drigalenko EI, Moy W, Freda J, et al. Transcriptome study of differential expression in schizophrenia. Hum Mol Genet 2013;22:5001–5014.
Tylee DS, Espinoza AJ, Hess JL, Tahir MA, McCoy SY, Rim JK, et al. RNA sequencing of transformed lymphoblastoid cells from siblings discordant for autism spectrum disorders reveals transcriptomic and functional alterations: evidence for sex-specific effects. Autism Res 2017;10:439–455.
Jong S, de, Boks MPM, Fuller TF, Strengman E, Janson E, Kovel CGFde, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLOS One 2012;7:e39498.
Kos MZ, Duan J, Sanders AR, Blondell L, Drigalenko EI, Carless MA, et al. Dopamine perturbation of gene co-expression networks reveals differential response in schizophrenia for translational machinery. Transl Psychiatry 2018;8:278.
Lin P, Yao Y, Hofmeister R, Tsien RY, Farquhar MG. Overexpression of CALNUC (Nucleobindin) increases agonist and Thapsigargin releasable Ca2+ storage in the Golgi. J Cell Biol 1999;145:279–289.
Tsukumo Y, Tomida A, Kitahara O, Nakamura Y, Asada S, Mori K, et al. Nucleobindin 1 controls the unfolded protein response by inhibiting ATF6 activation. J Biol Chem 2007;282:29264–29272.
Tulke S, Williams P, Hellysaz A, Ilegems E, Wendel M, Broberger C. Nucleobindin 1 (NUCB1) is a Golgi-resident marker of neurons. Neuroscience 2016;314:179–188.
Sundarrajan L, Blanco AM, Bertucci JI, Ramesh N, Canosa LF, Unniappan S. Nesfatin-1-like peptide encoded in nucleobindin-1 in goldfish is a novel anorexigen modulated by sex steroids, macronutrients and daily rhythm. Sci Rep 2016;6:28377.
Schalla MA, Stengel A. Current understanding of the role of Nesfatin-1. J Endocr Soc 2018;2:1188–1206.
Bloem B, Xu L, Morava E, Faludi G, Palkovits M, Roubos EW, et al. Sex-specific differences in the dynamics of cocaine- and amphetamine-regulated transcript and nesfatin-1 expressions in the midbrain of depressed suicide victims vs. controls. Neuropharmacology 2012;62:297–303.
Hofmann T, Elbelt U, Ahnis A, Rose M, Klapp BF, Stengel A. Sex-specific regulation of NUCB2/nesfatin-1: differential implication in anxiety in obese men and women. Psychoneuroendocrinology 2015;60:130–137.
Xiao M-M, Li J-B, Jiang L-L, Shao H, Wang B-L. Plasma nesfatin-1 level is associated with severity of depression in Chinese depressive patients. BMC Psychiatry 2018;18.
Sönnichsen B, Lowe M, Levine T, Jämsä E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol 1998;140:1013–1021.
Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic Cph Den 2007;8:758–773.
Young KG, Kothary R. Dystonin/Bpag1 is a necessary endoplasmic reticulum/nuclear envelope protein in sensory neurons. Exp Cell Res 2008;314:2750–2761.
Ryan SD, Ferrier A, Sato T, O’Meara RW, De Repentigny Y, Jiang SX, et al. Neuronal dystonin isoform 2 is a mediator of endoplasmic reticulum structure and function. Mol Biol Cell 2012;23:553–566.
Welihinda AA, Tirasophon W, Kaufman RJ. The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr 2018;7:293–300.
Carreras-Sureda A, Pihán P, Hetz C. Calcium signaling at the endoplasmic reticulum: fine-tuning stress responses. Cell Calcium 2018;70:24–31.
Bixo M, Johansson M, Timby E, Michalski L, Bäckström T. Effects of GABA active steroids in the female brain with a focus on the premenstrual dysphoric disorder. J Neuroendocrinol. 2018;30.
Van Wingen GA, Van Broekhoven F, Verkes RJ, Petersson KM, Bäckström T, Buitelaar JK, et al. Progesterone selectively increases amygdala reactivity in women. Mol Psychiatry 2008;13:325.
Mansour I, Reznikoff-Etievant MF, Netter A. No evidence for the expression of the progesterone receptor on peripheral blood lymphocytes during pregnancy. Hum Reprod Oxf Engl 1994;9:1546–1549.
Scariano JK, Emery-Cohen AJ, Pickett GG, Morgan M, Simons PC, Alba F. Estrogen receptors alpha (ESR1) and beta (ESR2) are expressed in circulating human lymphocytes. J Recept Signal Transduct Res 2008;28:285–293.
Dosiou C, Hamilton AE, Pang Y, Overgaard MT, Tulac S, Dong J, et al. Expression of membrane progesterone receptors on human T lymphocytes and Jurkat cells and activation of G-proteins by progesterone. J Endocrinol 2008;196:67–77.
Pierdominici M, Maselli A, Colasanti T, Giammarioli AM, Delunardo F, Vacirca D, et al. Estrogen receptor profiles in human peripheral blood lymphocytes. Immunol Lett 2010;132:79–85.
Gross KS, Mermelstein PG. Estrogen receptor signaling through metabotropic glutamate receptors. Vitam Horm 2020;114:211–232.
Srivastava DP, Waters EM, Mermelstein PG, Kramár EA, Shors TJ, Liu F. Rapid estrogen signaling in the brain: implications for the fine-tuning of neuronal circuitry. J Soc Neurosci 2011;31:16056–16063.
Bali N, Arimoto JM, Iwata N, Lin SW, Zhao L, Brinton RD, et al. Differential responses of progesterone receptor membrane component-1 (Pgrmc1) and the classical progesterone receptor (Pgr) to 17β-estradiol and progesterone in hippocampal subregions that support synaptic remodeling and neurogenesis. Endocrinology 2012;153:759–769.
Petersen SL, Intlekofer KA, Moura-Conlon PJ, Brewer DN, Del Pino Sans J, Lopez JA. Novel progesterone receptors: neural localization and possible functions. Front Neurosci 2013;7:164.
So J, Warsh JJ, Li PP. Impaired endoplasmic reticulum stress response in B-lymphoblasts from patients with bipolar-I disorder. Biol Psychiatry 2007;62:141–147.
Hayashi A, Kasahara T, Kametani M, Toyota T, Yoshikawa T, Kato T. Aberrant endoplasmic reticulum stress response in lymphoblastoid cells from patients with bipolar disorder. Int J Neuropsychopharmacol 2009;12:33–43.
Pfaffenseller B, Wollenhaupt-Aguiar B, Fries GR, Colpo GD, Burque RK, Bristot G, et al. Impaired endoplasmic reticulum stress response in bipolar disorder: cellular evidence of illness progression. Int J Neuropsychopharmacol 2014;17:1453–1463.
Hendrick V, Altshuler LL. Recurrent mood shifts of premenstrual dysphoric disorder can be mistaken for rapid-cycling bipolar II disorder. J Clin Psychiatry 1998;59:479–480.
Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011;469:343–349.
Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006;439:871.
Höglinger GU, Melhem NM, Dickson DW, Sleiman PMA, Wang L-S, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet 2011;43:699–705.
Latourelle JC, Pankratz N, Dumitriu A, Wilk JB, Goldwurm S, Pezzoli G, et al. Genomewide association study for onset age in Parkinson disease. BMC Med Genet 2009;10:98.
Andruska N, Zheng X, Yang X, Helferich WG, Shapiro DJ. Anticipatory estrogen activation of the unfolded protein response is linked to cell proliferation and poor survival in estrogen receptor α-positive breast cancer. Oncogene 2015;34:3760–3769.
Zheng X, Andruska N, Yu L, Mao C, Kim JE, Livezey M, et al. Interplay between steroid hormone activation of the unfolded protein response and nuclear receptor action. Steroids 2016;114:2–6.
Nilsen J, Diaz Brinton R. Mechanism of estrogen-mediated neuroprotection: regulation of mitochondrial calcium and Bcl-2 expression. Proc Natl Acad Sci USA 2003;100:2842–2847.
Bäckström T, Bixo M, Johansson M, Nyberg S, Ossewaarde L, Ragagnin G, et al. Allopregnanolone and mood disorders. Prog Neurobiol 2014;113:88–94.
Wohleb ES, Franklin T, Iwata M, Duman RS. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci. 2016;17:497–511.
Brites D, Fernandes A. Neuroinflammation and depression: microglia activation, extracellular microvesicles and microRNA dysregulation. Front Cell Neurosci. 2015;9:476.
Petschner P, Gonda X, Baksa D, Eszlari N, Trivaks M, Juhasz G, et al. Genes linking mitochondrial function, cognitive impairment and depression are associated with endophenotypes serving precision medicine. Neuroscience 2018;370:207–217.
Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008;454:455–462.
McEwen BS, Milner TA. Understanding the broad influence of sex hormones and sex differences in the brain. J Neurosci Res 2017;95:24–39.
Brinton RD. The healthy cell bias of estrogen action: mitochondrial bioenergetics and neurological implications. Trends Neurosci. 2008;31:529–537.
Gold PW, Licinio J, Pavlatou MG. Pathological parainflammation and endoplasmic reticulum stress in depression: potential translational targets through the CNS insulin, klotho and PPAR-γ systems. Mol Psychiatry 2013;18:154–165.
Levy NA, Janicak PG. Calcium channel antagonists for the treatment of bipolar disorder. Bipolar Disord 2000;2:108–119.
Pollack MH, Rosenbaum JF, Hyman SE. Calcium channel blockers in psychiatry: potential for a growing role in pharmacotherapy. Psychosomatics 1987;28:356–369.
Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci. Res. 2009;87:1953–1959.
Cai C, Langfelder P, Fuller TF, Oldham MC, Luo R, van den Berg LH, et al. Is human blood a good surrogate for brain tissue in transcriptional studies? BMC Genomics 2010;11:589.
Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry 2004;28:559–576.
Acknowledgements
We thank Cheryl Marietta and Longina Akhtar for technical assistance. This work was written as part of P.J.S. and D.G.’s official duties as Government employees. Views expressed here do not represent the views of the NIMH, NIH, HHS, or the United States Government. This research was supported by the Intramural Research Program of the NIMH, NIH. NIMH Protocols 90-M-0088 NCT0001259, 92-M-0174 NCT0001322; Projects #MH002865, #AA000301.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
DRR serves on the Clinical Advisory Board of Sage Therapeutics, and reports consulting fees and travel reimbursement from Sage Therapeutics.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, H.J., Goff, A., Rudzinskas, S.A. et al. Altered estradiol-dependent cellular Ca2+ homeostasis and endoplasmic reticulum stress response in Premenstrual Dysphoric Disorder. Mol Psychiatry 26, 6963–6974 (2021). https://doi.org/10.1038/s41380-021-01144-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01144-8
This article is cited by
-
Neuroactive steroid hormone trajectories across the menstrual cycle in premenstrual dysphoric disorder (PMDD): the PHASE study
Molecular Psychiatry (2024)
-
Intrinsically dysregulated cellular stress signaling genes and gene networks in postpartum depression
Molecular Psychiatry (2023)
-
GABA-ergic Modulators: New Therapeutic Approaches to Premenstrual Dysphoric Disorder
CNS Drugs (2023)
-
In search of sex-related mediators of affective illness
Biology of Sex Differences (2021)