Abstract
Renal cell carcinomas with t(6;11) chromosome translocation has been classically characterized by the rearrangement of the TFEB gene, located on chromosome 6, and MALAT1 gene, located on chromosome 11. Recently, a few other genes have been described as fusion partners in TFEB rearranged renal cell carcinomas. Although most of TFEB rearranged renal cell carcinomas have an indolent behavior, in the rare cases of advanced metastatic disease targeted therapy and predictive markers remain lacking. In the present study, we collected 13 TFEB rearranged renal cell carcinomas, confirmed by FISH, analyzing their morphology and exploring the novel gene partners. Looking for predictive markers, we have also performed PDL1 immunohistochemical analysis by using four different assays (E1L3N, 22C3, SP142, and SP263). MALAT1 gene rearrangement has been found in ten tumors, five cases showing classical biphasic morphology with “rosettes”, five cases without “rosettes” mimicking other renal cell carcinomas or epithelioid angiomyolipoma/pure epithelioid PEComa. We identified two different partner genes, ACTB and NEAT1, the latter previously unreported and occurring in a tumor with an unusual solid and cystic appearance. In both cases, the “rosettes” were absent. In one case no gene partner was identified. Overall, in 12 of 13 TFEB-rearranged renal cell carcinomas staining for PDL1 SP263 was observed, whereas the other antibodies were less reliable or more difficult to interpret. In conclusion, we described the third case of ACTB-TFEB rearranged renal cell carcinoma and a novel NEAT1-TFEB rearranged renal cell carcinoma, both without the distinctive biphasic morphology typical of t(6;11) renal cell carcinoma. Finally, PDL1 SP263 was constantly expressed in TFEB rearranged renal cell carcinoma with possible clinical benefit which requires further investigations.
Similar content being viewed by others
Introduction
t(6;11) renal cell carcinoma is classified as a subtype of the MiT family renal cell carcinoma which also includes the more common Xp11 translocation renal cell carcinoma, harboring TFEB and TFE3 gene fusions, respectively [1]. Both were initially described in the pediatric population, although it has been demonstrated that these neoplasms can occur in adults as well [2,3,4]. Over the last years, several manuscripts have reported a broad range of morphology of Xp11 translocation renal cell carcinoma and the several partner genes involved in the translocation with TFE3 gene [5,6,7]. A distinctive biphasic morphology with larger epithelioid cells and smaller cells clustered around eosinophilic spheres and MALAT1/TFEB rearrangement has been consistently shown in t(6;11) renal cell carcinoma [4]. However, recently, a variety of morphologies has also been described in t(6;11) renal cell carcinoma. In addition to the classic biphasic histopathologic features, cases mimicking epithelioid angiomyolipoma/pure epithelioid PEComa or resembling other renal cell carcinomas have been reported [8,9,10]. Along with the expanding morphological spectrum, an expanding genomic spectrum has also been recently demonstrated in which the TFEB transcription factor gene is translocated to other genes such as KHDRBS2, CADM2, COL21A1, ACTB, EWSR1, and CLTC [11,12,13,14].
While aggressive behavior is frequently observed (47%) in Xp11 renal cell carcinoma and therapeutic options and predictive markers has been proposed, t(6;11) renal cell carcinoma has an indolent clinical course with an aggressive behavior observed in roughly 17% of cases. Targeted therapy and predictive markers remain lacking in t(6;11) tumors [15]. Interestingly, programmed cell death ligand 1 (PDL1), also known as cluster of differentiation 274 (CD274), immunohistochemical expression (clone E1L3N) has been observed in the only two cases tested of t(6;11) renal cell carcinoma raising the possibility of a potential immunotherapeutic approach in the future [16].
In the study, we sought to comprehensively evaluate 13 cases of renal cell carcinoma with TFEB translocation analyzing their morphology and exploring the novel gene partners. In addition, we have performed PDL1 immunohistochemical analysis by using four different assays.
Material and methods
Patients and samples
Thirteen renal cell carcinomas with TFEB gene translocation were retrieved from the files of Department of Pathology University of Verona and Pederzoli Hospital, Peschiera del Garda, Verona. Five tumors were in-house cases; eight tumors were consultant cases. Eight cases have been previously reported [8, 17] and five unpublished cases have been added. The number of blocks from which hematoxylin eosin-stained sections were available for each tumor ranged from 1 to 42 (mean 18; median 16), six tumors were entirely submitted for microscopic evaluation. All slides were reviewed by two authors (AC, GM). For each case, the following morphologic features were recorded: solid, nested, tubulocystic, and papillary architecture, the presence or absence of pseudocapsule, necrosis, and psammoma bodies. Regarding cellular features, the presence of a small cell component around basement membrane-like material (“rosettes”), eosinophilic and clear cytoplasm, and nucleolar grade according to ISUP/WHO 2016 were assessed.
Immunohistochemistry
Representative sections from tissue blocks of renal cell carcinomas with TFEB translocation were immunohistochemically stained for the following antibodies: PAX8 (clone BC12, DSB), Cathepsin K (clone 3F9, dilution 1:2000, Abcam), HMB45 (dilution 1:30, Dako), Melan-A (clone A103, dilution 1:50, Novocastra), CD68 (clone PG-M1, dilution 1:50, Dako), cytokeratin 7 (clone RN7, dilution 1:100, Novocastra), CD10 (clone 56C6, dilution 1:50, Novocastra), alpha-methylacyl-CoA racemase AMACR (clone 13H7, dilution 1:25, Dako) and carbonic anhydrase 9 (polyclonal rabbit, dilution 1:1000, Abcam). All samples were processed using a sensitive “Bond polymer Refine” detection system in an automated Bond immunohistochemistry instrument (Leica Biosystems).The most commonly used immunohistochemical assays developed to determine PDL1 expression level were performed: SP263 and SP142 on the Ventana Benchmark Ultra platform according to the manufacturer’s instruction, while staining for 22C3 (monoclonal mouse anti-human PDL1, Dako) was performed following general instructions for immunohistochemical staining Dako, and E1L3N (dilution 1:500, Cell Signaling) was carried out on an automated Bond immunohistochemistry instrument (Leica Biosystems). The appropriate positive (tonsil and placenta) and negative controls were concurrently carried out. Labeling for each marker was recorded as the percentage of positive neoplastic cells.
Fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization (FISH) was carried out in all t(6;11) renal cell carcinoma using dual-color break apart TFEB probe (Cytotest Inc, Rockville, MD 20850, USA) as previously described [8]. Briefly, 3 µm sections were cut from formalin-fixed paraffin-embedded (FFPE) tissue blocks and mounted on positively charged slides. The slides were dried for 1 h at 60 °C then deparaffinized, rehydrated, and fixed in methanol/acetic acid 3:1 for 5 min. Pretreatment was performed at 85 °C for 30 min with 0,1 citrate buffer (pH6) solution followed by pepsin (4 mg/ml in 0.9% NaCl, pH 1,5) treatment for 8 min at 37 °C. After washing and dehydration, 10 µl probe was applied tothe selected area and sealed with rubber cement. Denaturation was assessed by incubating the slides at 80 °C for 10 min in a humidified atmosphere (Thermobrite System) followed by hybridization overnight at 37 °C. The rubber cement and the coverslip were removed and the slides were washed in 2X SSC/0,3% NP40 for 15 min at room temperature and then at 72 °C for 2 min. Next, the tissue sections were counterstained with DAPI antifade (Prolong Gold Antifade Reagent Life Technologies) and examined under an X60- X100 oil immersion objective using an Olympus BX61 fluorescence microscope equipped with filters that visualize the different wavelengths of the fluorescent probe.
Scoring was performed by two experienced pathologists (AC and MB). At least 100 neoplastic non overlapping nuclei were included in the scoring.
Molecular analysis
Total nucleic acid was extracted from sections from FFPE patient tissue samples using truXTRAC FFPE total NA Plus kit (Covaris, Inc. Woburn, MA). After measured RNA quantity using Qubit fluorometer (Life Technologies, Carlsbad, CA), the target enriched cDNA libraries were prepared with a custom designed Archer FusionPlex panel with 94 target genes (ArcherDX, Inc. Boulder, CO) as per manufacture’s instruction. The assay utilizes Anchored Multiplex PCR (AMP) technology, which utilizes unidirectional gene-specific primers (GPSs) that enrich both known and unknown fusion gene partners. All libraries were purified and sequenced on the Illumina NextSeq instrument (Illumina, San Diego, CA). Gene fusion events were analyzed using Archer Analysis software (version 6.2).
Results
Thirteen cases of renal cell carcinoma with TFEB gene rearrangements confirmed by FISH were analyzed for fusion partners. In one case no gene partner was identified. The results for specific subtypes are presented according to the specific gene rearrangement identified. The clinicopathological, molecular, and immunohistochemical results were summarized in Tables 1 and 2.
MALAT1-TFEB renal cell carcinoma
MALAT1-TFEB renal cell carcinoma with “rosettes”
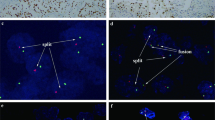
The MALAT1-TFEB fusion pattern was identified in five cases. The distinctive biphasic morphology with larger epithelioid cells and smaller cells with dark nuclei clustering around hyaline material in a background of sheets mainly composed by epithelioid clear cells was observed in five tumors. Overall, “rosettes” were identified in ~60% of the slides available for those tumors (61 of 104 slides for 5 tumors). In case #5, the biphasic morphology present in the original primary tumor, was absent in the renal recurrence observed 2 years later (17 slides examined for a 10 cm in diameter mass), morphologically resembling clear cell renal cell carcinoma with extensive necrosis. Calcifications were variably identified. Morphologically, no significant immune cells infiltrate was seen, except in one case (case #3) in which small perivascular immune cells infiltrates were observed. Both melanogenesis markers (HMB45 and Melan-A) and cathepsin K were observed in those tumors. The neoplastic cells were usually negative for PDL1 SP142, staining was faint with a variable percentage of positivity for 22C3 and E1L3N, whereas strong PDL1 SP263 was observed in all cases. Immune cells, when present, were negative for PDL1, whereas the scattered macrophages, present in all cases, were PDL1 positive. The frequency of split TFEB fluorescent signals by FISH was high (mean 74% and median 75%) (Figs. 1 and 4).
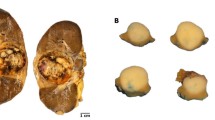
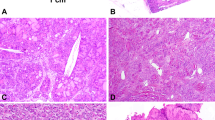
The macroscopic appearance of case # 7 (A), morphologically characterized by numerous “rosettes” (B), and lymph-node metastasis (C). The lack of “rosettes” of case #8 demonstrating sheets of large epithelioid cells with deeply eosinophilic cytoplasm resembling epithelioid angiomyolipoma/pure epithelioid PEComa (D). All MALAT1-TFEB rearranged renal cell carcinomas showed a strong and diffuse expression of cathepsin K (E) and consistent PDL1 SP263 positivity (F).
MALAT1-TFEB renal cell carcinoma without “rosettes”
Those tumors (five cases) were mainly characterized by a solid-alveolar architecture made up of medium-large epithelioid cells with abundant clear to eosinophilic cytoplasm and prominent nucleoli resembling epithelioid angiomyolipoma/pure epithelioid PEComa. The presence of eosinophilic cells was a common feature ranging from an isolated-small clusters of cells to a predominant component. Calcifications and necrosis were variably present. On hematoxylin-eosin slides, no significant immune cells infiltrate was identified. Both melanogenesis markers (HMB45 and Melan-A) and cathepsin K was seen in those tumors. Among those cases, one tumor was completely negative for all the PDL1 antibodies maybe due to the preanalytical phase whereas the others variably expressed PDL1. E1L3N tended to be weaker and in general more difficult to interpret. Higher PDL1 expression, mainly in terms of intensity, was detected by using SP263. Scattered macrophages, present in all cases, were PDL1 positive. The frequency of split TFEB fluorescent signals by FISH was high (mean 77% and median 78%) (Figs. 1 and 4).
ACTB-TFEB renal cell carcinoma
The ACTB-TFEB fusion pattern was identified in one case. The tumor was a solid mass of 6.5 cm in the greatest dimension, brown at cut surface with hemorrhagic areas. Microscopically, the neoplasm was made up of medium-sized polygonal clear to eosinophilic cells with conspicuous nucleoli (G3 by ISUP/WHO 2016) arranged in a solid-alveolar pattern, resembling epithelioid angiomyolipoma/pure epithelioid PEComa. Hemosiderin-laden macrophages were observed in the tumor. Focal bone metaplasia was identified. Neither calcification nor necrosis was seen. Mitotic figures were occasionally encountered (0–1 per 10 HPF). Smaller lymphocyte-like cells grouped around collagenous spherules formed by basement membrane material was not present in any of the six slides available. A single small aggregate of lymphocytes was found. Immunolabeling for Melan-A and HMB45 was easily found within the tumor whereas cathepsin K expression, initially considered negative, was focal and extremely weak. Membrane staining for PDL1 was observed by using three different antibodies (22C3, SP263, E1L3N) with the same percentage of neoplastic positive cells and negative by using SP142; the staining with SP263 was the most intense. The few lymphocytes identified were PDL1 negative. All antibodies of PDL1 labeled hemosiderin-laden macrophages. The frequency of split TFEB fluorescent signals by FISH was high (91%) in which the distance of red and green signals was greater than twice signal diameter (Figs. 2 and 4).
A solid mass arising in the renal parenchyma (A) characterized by medium-sized clear to eosinophilic cells arranged in nestswith hemosiderin-laden macrophages (B, C). The tumor cells were positive for HMB45 (D) and PDL1 SP263 (E). FISH assays demonstrated the TFEB gene rearrangement in 91% of the neoplastic cells (F).
NEAT1-TFEB renal cell carcinoma
The NEAT1-TFEB fusion pattern was identified in one case. Grossly, the tumor was 3 cm in diameter displaying a multicystic and solid appearance. Morphologically, it was a well-demarcated solid and cystic mass with a fibrous pseudocapsule. The cells lining the cystic septa showed abundant vacuolated clear and eosinophilic cytoplasm with round nuclei and pinpoint nucleoli (G2 by ISUP/WHO 2016). Melanin pigment and psammoma bodies were broadly present. Mitotic figures were occasionally found (0–1 per 10 HPF). No necrosis was observed. The classic biphasic morphology with “rosettes” was not seen in the completely embedded tumor. Histologically, no significant immune cells infiltrate was observed. The neoplastic cells stained diffusely for cathepsin K and expressed both melanogenesis markers (HMB45 and Melan-A) in variable percentages. All antibodies of PDL1 labeled the tumor with different percentage and intensity and the numerous macrophages present in the cystic component. There was a relevant increment in terms of intensity of cell positivity from SP263 on Ventana platform. The frequency of split TFEB fluorescent signals by FISH was high (65%) in which the distance of red and green signals was greater than twice signal diameter (Figs. 3 and 4).
Schematic representation of RNA sequencing results showing TFEB fusion transcripts with MALAT1 (A), ACTB (B), and NEAT1 (C). Exons of TFEB and the sequencing near the breakpoint are shown in red and exons of its partner gene and the sequencing near the breakpoint are shown in blue. Arrows indicate breakpoint and its chromosomal locations are listed below.
Discussion
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is the most common fusion partner gene in TFEB rearranged renal cell carcinoma [18, 19]. A recent manuscript has drawn the attention to the different fusion variants on TFEB rearranged renal cell carcinoma and the possible correlations with specific clinicopathologic features [12]. In the present study, we described the third case of ACTB-TFEB renal cell carcinoma and the novel finding of the Nuclear Enriched Abundant Transcript 1 (NEAT1) fusion partner in NEAT1-TFEB renal cell carcinoma providing its first morphological characterization. The NEAT1 gene, located at 11q13.1, the same locus of MALAT1, produces a functionally conserved long non-coding RNA reported to be frequently deregulated in various types of cancers [20]. NEAT1 plays an indispensable role in the formation and integrity of specific nuclear structures called paraspeckles, which are membranelles nuclear bodies [21]. Interestingly, the MALAT1 gene is also known as NEAT2 (nuclear-enriched abundant transcript 2) [22] and produces a long non-coding RNA localized at nuclear domains known as nuclear speckles [23]. NEAT1 is a new partner gene in TFEB rearranged renal cell carcinoma but it has been previously reported in Xp11 translocation renal cell carcinoma [24]. Of note, NEAT1-TFE3 renal cell carcinoma case displayed a biphasic morphology due to larger epithelioid cells and small lymphocyte-like cells, resembling a t(6;11) renal cell carcinoma, with focal melanin pigment. Even though the distinctive biphasic appearance has not been seen in any slides of NEAT1-TFEB renal cell carcinoma, the occurrence of melanin pigment was a characteristic feature that has not been previously reported in not-MALAT1 TFEB rearranged renal cell carcinomas in the literature.
Actin Beta (ACTB) gene is located on chromosome 7p22.1 and encodes one of six different actin proteins. Two previously reported ACTB-TFEB renal cell carcinoma showed two different morphologies, one case mimicking epithelioid angiomyolipoma/pure epithelioid PEComa, as the present case, the other demonstrated solid and papillary architecture [12]. From an immunohistochemical point of view, the present case of ACTB-TFEB renal cell carcinoma demonstrated a very focal and weak expression of cathepsin K, in contrast to other cases of TFEB rearranged renal cell carcinoma [25]. Unfortunately, the immunohistochemical profile has not been reported in the other two cases of ACTB-TFEB renal cell carcinoma and it is difficult to provide if this feature is characteristic of the tumor with this kind of rearrangement. As detailed in Table 3, among not-MALAT1 TFEB rearranged renal cell carcinoma, ACTB is the most frequent reported. Like other fusion partners of TFEB (e.g., CADM2, EWSR1, and CLTC), ACTB gene is not located on chromosome 11 further supporting the rational for using the terminology of TFEB rearranged renal cell carcinoma for this group of neoplasms rather than the t(6;11) renal cell carcinoma nomenclature.
The present morphologic data of MALAT1-TFEB renal cell carcinoma highlights two different subsets: i) a subset with “rosettes” in which the diagnosis is usually straightforward but when the biphasic appearance is focal can be confused mainly with clear cell renal cell carcinoma and Xp11 translocation renal cell carcinoma; ii) a subset lacking “rosettes” despite extensive sampling or complete embedding. In this subset the lesions are composed of eosinophilic cells where the challenging differential diagnosis is epithelioid angiomyolipoma/pure epithelioid PEComa. It should be noted that the “rosettes”, when present, are easily found in the current series, hence a standard sampling of the mass should allow their recognition. Herein, the typical biphasic morphology is encountered in approximately 40% (5/13) of the overall series of TFEB rearranged renal cell carcinoma, and 50% (5/10) among MALAT1-TFEB renal cell carcinomas. A similar percentage, 41% (9/22), has been reported by Gupta et al. in TFEB rearranged renal cell carcinomas and 44% (8/18) among MALAT1-TFEB renal cell carcinomas [26]. A lower percentage, 29% (9/31) has been found by Xia et al. in TFEB rearranged renal cell carcinomas and 41% (9/22) among MALAT1-TFEB renal cell carcinomas [12]. The higher percentage described in the present study may be explained by the great number of slides reviewed for each case. Necrosis has been found in five cases, four of them with aggressive clinical course, either with or without “rosettes”. The only case with necrosis (case # 6) harboring an apparently indolent behavior has a very short follow up (6 months). Xia et al. described two cases with necrosis, both aggressive, in tumors with or without the distinctive biphasic appearance [12]; whereas Gupta and coauthors found the necrosis in four cases, only in tumors without the biphasic features, and a single case with metastasis (the others with no available follow-up) [26]. These findings support the correlation of necrosis with tumor aggressiveness, as previously suggested in TFEB rearranged renal cell carcinoma [12] and Xp11 translocation renal cell carcinoma [27].
TFEB rearranged renal cell carcinoma may present with advanced metastatic disease. Given the increasing utilization of immunotherapy in advanced renal cell carcinoma, we investigated PDL1 expression in those tumors. A recent study described only 2 cases of TFEB rearranged renal cell carcinoma positive for PDL1 (clone E1L3N). Taking into consideration the possibility of inter-assay variation in PDL1 assessment in other solid tumors, we performed PDL1 immunohistochemical analysis using four different assays. All cases but one stained for PDL1 with variable intensity and percentage of positive neoplastic cells. Among these, SP142 seemed to be less reliable, E1L3N and 22C3 tended to be weaker and in general more difficult to interpret, whereas SP263 was the most robust antibody. The different level of PDL1 staining between assays is in part explained by the epitope binding variance among antibodies and more likely attributable to assay or platform variables [28]. The almost universal immunolabelling for PDL1 in our cases support immunotherapy as a therapeutic option in patients with those tumors. However, further clinical studies should be conducted to evaluate the efficacy of this treatment in the patients with advanced TFEB rearranged renal cell carcinomas. Moreover, the underlying mechanism of PDL1 expression is incompletely understood. A possible hypothesis is that TFEB translocation produces immunogenic neoantigen that induces a T cell response. Another fascinating explanation is that PDL1 overexpression is driven by TFEB which directly regulates PDL1, binding PDL1 promoter [29]. In this scenario, one would expect that PDL1 is more expressed in cases in which TFEB is more present. It has been demonstrated that higher levels of TFEB gene expression is correlated with biphasic morphology [26], however, we did not observe any differences of PDL1 labeling among the tumors with or without “rosettes”.
In summary, we described the third case of ACTB-TFEB rearranged renal cell carcinoma and a novel NEAT1-TFEB rearranged renal cell carcinoma, both without the distinctive biphasic morphology typical of t(6;11) renal cell carcinoma. In our series, this morphological feature is present in 40% of TFEB rearranged renal cell carcinomas, and 50% of MALAT1-TFEB renal cell carcinomas, even with a standard sampling of the tumor. Finally, PDL1 (clone SP263) is constantly expressed in TFEB rearranged renal cell carcinoma with possible clinical benefit which required further investigations.
References
Moch H, Humprey PA, Ulbright TM, Reuter VE. WHO classification of tumours of the urinary system and male genital organs. Lyon, France: Cancer IARC; 2016.
Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–92.
Argani P, Hawkins A, Griffin CA, Goldstein JD, Haas M, Beckwith JB, et al. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol. 2001;158:2089–96.
Argani P. MiT family translocation renal cell carcinoma. Semin Diagn Pathol. 2015;32:103–13.
Argani P, Zhong M, Reuter VE, Fallon JT, Epstein JI, Netto GJ, et al. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol. 2016;40:723–37.
Wang XT, Xia QY, Ye SB, Wang X, Li R, Fang R, et al. RNA sequencing of Xp11 translocation-associated cancers reveals novel gene fusions and distinctive clinicopathologic correlations. Mod Pathol. 2018;31:1346–60.
Hayes M, Peckova K, Martinek P, Hora M, Kalusova K, Straka L, et al. Molecular-genetic analysis is essential for accurate classification of renal carcinoma resembling Xp11.2 translocation carcinoma. Virchows Arch. 2015;466:313–22.
Caliò A, Brunelli M, Segala D, Pedron S, Tardanico R, Remo A, et al. t(6;11) renal cell carcinoma: a study of seven cases including two with aggressive behavior, and utility of CD68 (PG-M1) in the differential diagnosis with pure epithelioid PEComa/epithelioid angiomyolipoma. Mod Pathol. 2018;31:474–87.
Inamura K, Fujiwara M, Togashi Y, Nomura K, Mukai H, Fujii Y, et al. Diverse fusion patterns and heterogeneous clinicopathologic features of renal cell carcinoma with t(6;11) translocation. Am J Surg Pathol. 2012;36:35–42.
Argani P, Lae M, Hutchinson B, Reuter VE, Collins MH, Perentesis J, et al. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol. 2005;29:230–40.
Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res. 2014;20:4129–40.
Xia QY, Wang XT, Fang R, Wang Z, Zhao M, Chen H, et al. Clinicopathologic and molecular analysis of the TFEB fusion variant reveals new members of TFEB translocation renal cell carcinomas (RCCs): expanding the genomic spectrum. Am J Surg Pathol. 2020;44:477–89.
Durinck S, Stawiski EW, Pavia-Jimenez A, Modrusan Z, Kapur P, Jaiswal BS, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47:13–21.
Cancer Genome Atlas Research N, Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374:135–45.
Caliò A, Segala D, Munari E, Brunelli M, Martignoni G. MiT family translocation renal cell carcinoma: from the early descriptions to the current knowledge. Cancers. 2019;11:1110.
Walter B, Gil S, Naizhen X, Kruhlak MJ, Linehan WM, Srinivasan R, et al. Determination of the expression of PD-L1 in the morphologic spectrum of renal cell carcinoma. J Cancer. 2020;11:3596–603.
Caliò A, Brunelli M, Segala D, Pedron S, Doglioni C, Argani P, et al. VEGFA amplification/increased gene copy number and VEGFA mRNA expression in renal cell carcinoma with TFEB gene alterations. Mod Pathol. 2019;32:258–68.
Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, et al. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet. 2003;12:1661–9.
Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci USA. 2003;100:6051–6.
Dong P, Xiong Y, Yue J, Hanley SJB, Kobayashi N, Todo Y, et al. Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front Genet. 2018;9:471.
Taiana E, Ronchetti D, Todoerti K, Nobili L, Tassone P, Amodio N, et al. LncRNA NEAT1 in paraspeckles: a structural scaffold for cellular DNA damage response systems? Noncoding RNA. 2020;6:26.
Seles M, Hutterer GC, Kiesslich T, Pummer K, Berindan-Neagoe I, Perakis S, et al. Current insights into long non-coding RNAs in renal cell carcinoma. Int J Mol Sci. 2016;17:573.
Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–14.
Pei J, Cooper H, Flieder DB, Talarchek JN, Al-Saleem T, Uzzo RG, et al. NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, new members of MiT family translocation renal cell carcinoma. Mod Pathol. 2019;32:710–6.
Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–22.
Gupta S, Argani P, Jungbluth AA, Chen YB, Tickoo SK, Fine SW, et al. TFEB expression profiling in renal cell carcinomas: clinicopathologic correlations. Am J Surg Pathol. 2019;43:1445–61.
Caliò A, Brunelli M, Segala D, Pedron S, Remo A, Ammendola S, et al. Comprehensive analysis of 34 MiT family translocation renal cell carcinomas and review of the literature: investigating prognostic markers and therapy targets. Pathology. 2020;52:297–309.
Lawson NL, Dix CI, Scorer PW, Stubbs CJ, Wong E, Hutchinson L, et al. Mapping the binding sites of antibodies utilized in programmed cell death ligand-1 predictive immunohistochemical assays for use with immuno-oncology therapies. Mod Pathol. 2020;33:518–30.
Zhang C, Duan Y, Xia M, Dong Y, Chen Y, Zheng L, et al. TFEB mediates immune evasion and resistance to mTOR inhibition of renal cell carcinoma via induction of PD-L1. Clin Cancer Res. 2019;25:6827–38.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Caliò, A., Harada, S., Brunelli, M. et al. TFEB rearranged renal cell carcinoma. A clinicopathologic and molecular study of 13 cases. Tumors harboring MALAT1-TFEB, ACTB-TFEB, and the novel NEAT1-TFEB translocations constantly express PDL1. Mod Pathol 34, 842–850 (2021). https://doi.org/10.1038/s41379-020-00713-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-00713-6
This article is cited by
-
Genomic alterations and diagnosis of renal cancer
Virchows Archiv (2024)
-
Papillary Renal Cell Carcinoma: A Review of Prospective Clinical Trials
Current Treatment Options in Oncology (2023)
-
A primary rectal neoplasm with novel DDX5-TFEB fusion
Virchows Archiv (2022)
-
TFE3 and TFEB-rearranged renal cell carcinomas: an immunohistochemical panel to differentiate from common renal cell neoplasms
Virchows Archiv (2022)
-
Diagnostic utility of one-stop fusion gene panel to detect TFE3/TFEB gene rearrangement and amplification in renal cell carcinomas
Modern Pathology (2021)