Abstract
The hyalinizing trabecular adenoma/tumor is a rare and poorly characterized follicular-derived thyroid neoplasm recently shown to harbor recurrent PAX8–GLIS1 or PAX8–GLIS3 gene fusions. Here we sought to define the repertoire of genetic alterations of hyalinizing trabecular tumors, and whether PAX8–GLIS3 fusions are pathognomonic for hyalinizing trabecular tumors. A discovery series of eight hyalinizing trabecular tumors was subjected to RNA-sequencing (n = 8), whole-exome sequencing (n = 3) or targeted massively parallel sequencing (n = 5). No recurrent somatic mutations or copy number alterations were identified in hyalinizing trabecular tumor, whereas RNA-sequencing revealed the presence of a recurrent genetic rearrangement involving PAX8 (2q14.1) and GLIS3 (9p24.2) genes in all cases. In this in-frame fusion gene, which comprised exons 1–2 of PAX8 and exons 3–11 of GLIS3, GLIS3 is likely placed under the regulation of PAX8. Reverse transcription RT-PCR and/or fluorescence in situ hybridization analyses of a validation series of 26 hyalinizing trabecular tumors revealed that the PAX8–GLIS3 gene fusion was present in all hyalinizing trabecular tumors (100%). No GLIS1 rearrangements were identified. Conversely, no PAX8–GLIS3 gene fusions were detected in a cohort of 237 control thyroid neoplasms, including 15 trabecular thyroid lesions highly resembling hyalinizing trabecular tumor from a morphological standpoint, as well as trabecular/solid follicular adenomas, solid/trabecular variants of papillary carcinoma, and Hurthle cell adenomas or carcinomas. Our data provide evidence to suggest that the PAX8–GLIS3 fusion is pathognomonic for hyalinizing trabecular tumors, and that the presence of the PAX8–GLIS3 fusion in thyroid neoplasms may be used as an ancillary marker for the diagnosis of hyalinizing trabecular tumor, thereby avoiding overtreatment in case of misdiagnoses with apparently similar malignant tumors.
Similar content being viewed by others
Introduction
Hyalinizing trabecular adenoma/tumor is a rare follicular-derived, thyroglobulin-producing, thyroid neoplasm composed of large trabeculae of elongated or polygonal eosinophilic cells admixed with intratrabecular hyaline material of the basal membrane type. Hyalinizing trabecular tumors share many nuclear features of those of papillary thyroid carcinoma, namely nuclear clearing, grooves, pseudoinclusions, and irregular borders [1]. This tumor was described as a benign follicular tumor (a single malignant case is on record among 118 benign hyalinizing trabecular tumors reviewed some years ago) [2, 3] and was variably named, as either adenoma or tumor or neoplasm of uncertain malignant potential. No etiological or risk factors are known for hyalinizing trabecular tumor and a real understanding of its nature is missing [2]. An association with papillary carcinomas is reported in up to 5% of cases and hyalinizing trabecular tumor itself has been related to papillary thyroid carcinoma based on shared morphological features and RET gene alterations in a fraction of cases [4, 5]. Besides RET–PTC rearrangements, no other papillary thyroid carcinoma-related genetic abnormalities, such as BRAF mutations [6] or microRNA expression [7, 8], have been described in association with hyalinizing trabecular tumor. Similarly, no follicular tumor-related alterations were found, including RAS gene mutations or PAX8–PPARG rearrangements, the latter originally claimed to be specific of follicular carcinomas [9].
Very recently, Nikiforova et al. [10] have reported on the presence of gene fusions involving PAX8 and GLIS genes as pathognomonic genomic features underlying hyalinizing trabecular tumor. These fusions, in particular the PAX8–GLIS3 rearrangement, were identified in 100% of tumors with cardinal unequivocal morphological and immunophenotypical features of hyalinizing trabecular tumors. By performing a thorough genomic characterization of this entity, the same authors were also able to show that the hyalinizing trabecular tumor typically shows a quiet genome with a low number of somatic mutations and no significant DNA copy number alterations [10].
In this study, we provide a thorough independent validation of the presence of the PAX8–GLIS3 fusion in a large multi-institutional series of 34 hyalinizing trabecular tumors, as opposed to its absence in 237 control thyroid neoplasms that also included 15 trabecular tumors of follicular derivation highly resembling hyalinizing trabecular tumor from a morphological standpoint, thus posing a potential diagnostic interpretational issue in both histopathologic and fine needle aspiration cytology diagnostic practice.
Material and methods
Cases
After obtaining approval by the IRBs and local research ethics committees from the authors’ institutions, the pathology files of the authors’ institutions (Città della Salute Hospital and Mauriziano Hospital in Torino, Italy; San Luigi Hospital in Orbassano-Torino, Italy; Service of Clinical Pathology, Lausanne University Hospital, Switzerland; IRCCS in Reggio Emilia, Italy; Cleveland Clinic, Ohio, USA; Ottawa Hospital, Canada) were searched for a diagnosis of either “hyalinizing trabecular tumor” or “hyalinizing trabecular adenoma” of the thyroid. Samples were anonymized prior to the analyses. Representative hematoxylin & eosin (H&E) and Ki67/MIB1 stained slides of 38 hyalinizing trabecular tumors were centrally reviewed by two pathologists with a expertize in thyroid neoplasms (MV and MP). The diagnosis of hyalinizing trabecular tumor was based on the strict diagnostic criteria reported in the WHO classification of endocrine tumors of the thyroid [1], as well as in previous publications [11, 12]. Upon pathological review, 34 cases were unanimously diagnosed as hyalinizing trabecular tumor; four cases were excluded and reclassified as follicular adenomas of the trabecular or Hurthle cell type (three tumors: # 13T, 15T, and 23T) and as Hurthle cell carcinoma based on signs of vascular invasion in the remaining case (# 24T) (Fig. 1).
Schematic representation of the tissue samples and sequencing methods employed in this study. Depiction of the discovery and of the validation series of hyalinizing trabecular tumors of the thyroid, and the comprehensive series of other thyroid neoplasms here investigated, including 15 cases (*) in which the morphological features were highly reminiscent of a hyalinizing trabecular tumors. The sequencing analysis methods utilized in each cohort are detailed
Of the 34 hyalinizing trabecular tumors included in this study, four (HTT02, HTT06, HTT12, and HTT33) also contained a small, classical papillary carcinoma in the same thyroid lobe, but topographically independent from the hyalinizing trabecular tumor nodule. In addition, a separate series of 237 neoplasms that represented a comprehensive landscape of thyroid lesions were used as controls, and comprised 119 papillary carcinomas, 24 follicular carcinomas, 13 poorly differentiated carcinomas, 25 anaplastic carcinomas, 15 medullary carcinomas, 18 Hurthle cell tumors, and 23 follicular adenomas (Fig. 1). Within this series, 15 tumors showed morphological features highly reminiscent of a hyalinizing trabecular tumor, i.e., prominent solid/trabecular growth pattern and more or less extensive hyalinization. These were appropriately classified in different categories (i.e., follicular adenomas of trabecular/embryonal variant, solid/trabecular variant of papillary carcinomas, Hurthle cell adenomas, and Hurthle cell carcinoma).
First, eight hyalinizing trabecular tumors were subjected as a discovery series to RNA- and DNA-sequencing (Fig. 1). Subsequently, a validation series of 26 hyalinizing trabecular tumors were subjected to reverse transcription (RT)-PCR and fluorescence in situ hybridization analyses (Fig. 1).
Microdissection and nucleic acid extraction
Eight-micron-thick sections were cut from formalin-fixed paraffin-embedded blocks of the tumor and normal thyroid tissue, stained with nuclear fast red and microdissected under a stereomicroscope as previously described [13] to ensure >80% of tumor cells content and that the normal tissue was devoid of neoplastic cells. RNA and genomic DNA were extracted and quantified as previously described [14].
Immunohistochemistry
Four-micron-thick sections were cut from the representative formalin-fixed paraffin-embedded block of the 34 hyalinizing trabecular tumor and of the 15 tumors showed morphological features highly reminiscent of a hyalinizing trabecular tumor, as well as from tissue microarrays blocks containing the remaining 222 control thyroid neoplasms. Slides were processed for Ki67 immunohistochemistry with an automatic immunostainer (Ventana BenchMark AutoStainer, Ventana Medical Systems, Tucson, USA), with the exception of the primary antibody incubation step, which was manually performed at room temperature for 30 min using the Ki67 clone MIB1 (Dako/Agilent, Glostrup, Denmark) diluted 1/50, after antigen retrieval with the commercial Ventana CC1 solution for 36 min. The evaluation of Ki67 relied on a qualitative assessment of the staining pattern (membranous vs. nuclear).
Whole-exome and targeted massively parallel sequencing
Microdissected tumor and normal DNA samples from eight hyalinizing trabecular tumors (discovery set) were subjected to whole-exome sequencing (WES, n = 3) [15] and to Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) sequencing (n = 5; Fig. 1), which targets all exons and selected introns of 410 cancer genes, as previously described [16, 17] (Supplementary Table S1). Three additional cases from the validation set were also subjected to MSK-IMPACT (Fig. 1).
Analysis of sequencing data was performed as previously described [15, 17]. In brief, somatic single-nucleotide variants were detected using MuTect (v1.0) [18], and small insertion and deletions (indels) by Strelka (v2.0.15) [19], VarScan2 (v2.3.7) [20], Lancet (v1.0.0) [21], and Scalpel (v0.5.3) [22]. Single-nucleotide variants and indels for which the tumor mutant allele fraction was <5 times that of the matched normal mutant allele fraction were excluded. Single-nucleotide variants and indels found at >1% global minor allele frequency in ExAC [23], as well as common SNPs reported in dbSNP (build 150) were also excluded. Copy number alterations and loss of heterozygosity were defined using FACETS [24]. ABSOLUTE (v1.0.6) [25] was employed to determine the cancer cell fraction of each mutation, as previously described [15, 17]. A mutation was classified as clonal if its probability of being clonal was >50% or if the lower bound of the 95% confidence interval of its cancer cell fraction was >90%, as previously described [15, 17]. Mutations affecting hotspot codons were annotated according to Chang et al. [26].
RNA-sequencing and fusion gene identification
RNA-sequencing was performed on eight hyalinizing trabecular tumors (Fig. 1, Supplementary Table S1) using validated protocols [15] employed at MSKCC’s Integrated Genomics Operation (IGO). In brief, paired-end RNA-sequencing was performed with 2 × 50 bp cycles on an Illumina HiSeq2000. Read pairs supporting fusion transcripts were identified using INTEGRATE [27], deFuse [28], and FusionCatcher [29], as previously described [15, 30]. To account for alignment artifacts and normal transcriptional variants, we excluded fusion gene and read-through candidates that were also identified in a set of 297 normal samples from The Cancer Genome Atlas [31]. The remaining in-frame candidate fusion genes were annotated using OncoFuse [32] to define their likelihood of constituting potential driver fusion genes. The presence of the in-frame candidate fusion genes identified was also assessed across 33 cancer types in the tumor fusion gene data portal [33] that comprises a list of 20,231 fusion genes.
PAX8–GLIS3 fluorescence in situ hybridization analysis
PAX8–GLIS3 fusion gene assessment by fluorescence in situ hybridization was performed using validated protocols at MSKCC’s Molecular Cytogenetics Core, as previously described [34]. Three-color PAX8–GLIS3 probe was employed, consisting of bacterial artificial chromosomes mapping to 5′PAX8 (green), 3′PAX8 (red), and 3′GLIS3 (orange). At least ten images per tumor region were captured and at least 50 nonoverlapping interphase nuclei with well-delineated contours were analyzed for the presence of the PAX8–GLIS3 fusion. Hyalinizing trabecular tumors were considered positive for the PAX8–GLIS3 fusion gene if >15% of the cells displayed at least one 5′PAX8–3′GLIS3 fusion signal.
RT-PCR
Total RNA was reverse-transcribed using SuperScript Vilo Master Mix (Life Technologies; Thermo Fisher Scientific), according to the manufacturer’s instructions. PCR amplification of 10 ng of cDNA was performed using specific primer sets designed based on the known fusion genes and breakpoints (Supplementary Table S2). PCR fragments were purified (ExoSAP-IT, Affymetrix) and sequenced on an ABI 3730 capillary sequencer using the ABI BigDye Terminator chemistry (v3.1, Life Technologies) according to the manufacturer’s protocol. Sequences of the forward and reverse strands were analyzed using MacVector software (MacVector, Inc., Cary, NC, USA). All analyses were performed in duplicate.
Results
Histologic features
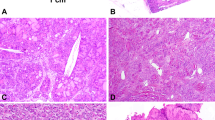
All 34 hyalinizing trabecular tumors included in this study displayed the cardinal morphological features of hyalinizing trabecular tumors: a growth pattern made of thick trabeculae; elongated or, more rarely, polygonal cells arranged with a perpendicular orientation compared to the trabecular axis; eosinophilic cytoplasm and clear nuclei with irregular borders, grooves, and pseudoinclusions; and finally the presence of more or less abundant hyaline material typically intratrabecular and often embedding single or groups of tumor cells (Fig. 2). In addition, all hyalinizing trabecular tumors displayed immunoreactivity at the cell membrane for the MIB1 monoclonal antibody to Ki67 (Fig. 2).
Representative micrographs of hyalinizing trabecular tumors of the thyroid. A 3-mm large well-circumscribed hyalinizing trabecular tumor (a) showing tumor cells arranged in trabeculae and small nests with abundant intratrabecular hyaline material (b), and another classical case of hyalinizing trabecular tumor (c) showing elongated and spindle cells with nuclear palisading and hyaline material in the background displaying the unique cell membrane reactivity with the MIB1 monoclonal antibody against Ki67 (d)
The 237 control cases, whose histology is detailed in Supplementary Table S3, had the expected morphology for each single tumor entity, with the exception of 15 selected cases that had a prominent solid and/or trabecular growth pattern and more or less extensive signs of hyalinization that resembled a hyalinizing trabecular tumor, but were eventually classified as variants of follicular adenoma (trabecular/embryonal variant, n = 8), Hurthle cell adenoma (solid/trabecular patterns, n = 2), papillary carcinoma (solid/trabecular variant, n = 3), Hurthle cell carcinoma (solid/trabecular patterns, n = 1), and poorly differentiated carcinoma (trabecular variant, n = 1) (Fig. 1). In all such cases, a hyalinizing trabecular tumor was originally suspected or diagnosed, based on the trabecular growth pattern and the presence of stromal hyaline material. The trabeculae were generally thin and merged, however, with compact solid nests made of cells haphazardly oriented along the axis of the cords (rather than perpendicularly) and with a deeply eosinophilic and granular cytoplasm, as commonly seen in Hurthle cells neoplasms. In addition, it is important to note that the variable degree of hyaline material observed in these cases was restricted to the space between the trabeculae and was not distributed inside them: indeed, a typical feature of hyalinizing trabecular tumors is represented by the tight contact of such material to the neoplastic cells. Finally, in all other thyroid neoplasms included the diagnosis of hyalinizing trabecular tumor was not supported due to the lack of the peculiar cell membrane reactivity pattern for Ki67/MIB1 (Fig. 3). Similarly, none of the other control thyroid neoplasms (n = 222) showed the peculiar cell membrane reactivity pattern for Ki67/MIB1.
Representative micrographs of thyroid neoplasms mimicking hyalinizing trabecular tumors. The figure illustrates a follicular adenoma of trabecular variant (a, 200×), a poorly differentiated thyroid carcinoma (b, 200×), and a Hurthle cell carcinoma (c, 200×) with prominent hyalinization, mimicking hyalinizing trabecular tumors on morphological ground. No membrane Ki67/MIB1 staining was observed, as exemplified here for the case of follicular thyroid adenoma with trabecular features (d, 200×)
Mutational landscape and copy number alterations in hyalinizing trabecular tumors
To assess the somatic genetic alterations, three and eight cases were subjected to WES (median depth of coverage of tumor 139× (range 137× to 164×) and normal 151× (range 131× to 161×) samples) and targeted MSK-IMPACT sequencing (median depth of coverage of tumor 311× (range 18× to 499×) and normal 174× (range 41× to 530×) samples), respectively (Supplementary Table S4).
Hyalinizing trabecular tumors displayed a low tumor burden, with a median of 16 (range 10–22) somatic mutations defined by WES, of which 11 (range 9–19) were nonsynonymous, while the MSK-IMPACT assay detected a median of 1 (range 1–2) somatic mutations of which 1 (range 1–2) was nonsynonymous. No recurrent somatic mutations were identified in the hyalinizing trabecular tumors analyzed. The majority of mutations were nonpathogenic missense mutations not affecting hotspot residues, and four hyalinizing trabecular tumors subjected to MSK-IMPACT sequencing did not harbor any mutations affecting the 410 cancer-related genes tested (Supplementary Figure S1). The potentially pathogenic nonsynonymous mutations identified included a subclonal FIP1L1 (p.L26R) missense mutation and a clonal ITK (p.R581W) missense mutation in HTT10 and HTT11, respectively. Analysis from MSK-IMPACT demonstrated a subclonal SPEN (p.P1892Qfs*6) frameshift mutation and a clonal KDM5A (p.R719H) missense mutation in HTT01, and a subclonal TP63 (p.X549) splice site mutation in HTT08 (Supplementary Fig. S1, Supplementary Table S5).
Copy number analysis revealed a diploid/near-diploid genome in the 11 hyalinizing trabecular tumors (Supplementary Fig. S2). No homozygous deletions or high-level amplifications were found in the hyalinizing trabecular tumors analyzed.
PAX8–GLIS3 fusion detection and validation in hyalinizing trabecular tumors
Based on the initial findings of a quiet genome and lack of recurrent pathogenic somatic mutations, the eight hyalinizing trabecular tumors of our discovery series were subjected to global RNA-sequencing analysis, which detected the presence of a recurrent, in-frame PAX8–GLIS3 fusion gene (8/8, 100%), resulting in a chimeric transcript composed of exons 1–2 of PAX8 and exons 3–11 of GLIS3 (Fig. 4a, Supplementary Table S2). The fusion of exons 1–2 of PAX8 to the 3′-end of GLIS3 results in the increased expression of exons 3–11 of GLIS3, as assessed using the RPKM counts (Fig. 4b). In the eight hyalinizing trabecular tumors and in a validation series of 26 hyalinizing trabecular tumors, the PAX8–GLIS3 fusion gene was confirmed/presented by fluorescence in situ hybridization (n = 12) and/or RT-PCR (n = 33) in all hyalinizing trabecular tumors (100%; Fig. 4c, d, Supplementary Table S3). Four hyalinizing trabecular tumors had a concurrent papillary carcinoma, and whilst the PAX8–GLIS3 fusion gene was present in the hyalinizing trabecular tumor, it was absent in the papillary carcinoma of these cases (Fig. 5).
Recurrent PAX8–GLIS3 fusion gene in hyalinizing trabecular tumors of the thyroid. a Schematic representation of the PAX8–GLIS3 fusion transcript including the exons and domains involved. The breakpoint of the 5′ and 3′ partner genes are represented as black vertical lines. Spanning reads are depicted and aligned to the predicted junction sequence. b Schematic representation the Reads Per Kilobase per Million (RPKM) mapped read counts of each exon of GLIS3. The GLIS3 fusion breakpoint is represented as a red dashed line. c Representative micrographs of fluorescent in situ hybridization for PAX8–GLIS3. d A representative Sanger sequencing electropherogram of the genomic PAX8–GLIS3 breakpoint (top) and frequency of PAX8–GLIS3 fusion gene in hyalinizing trabecular tumors involved in this study determined by RT-PCR (bottom). e Frequency of the PAX8–GLIS3 fusion gene in hyalinizing trabecular tumors and other thyroid neoplasms of the control cohort determined by RT-PCR. AC anaplastic carcinomas, FA follicular adenomas, FC follicular carcinomas, HCT hurthle cell tumors, HTT hylinizing trabecular tumors, MC medullary carcinomas, PDC poorly differentiated carcinomas, PTC papillary thyroid carcinomas. f Frequency of PAX8–GLIS3 fusion gene in hyalinizing trabecular tumors analyzed in this study and by Nikiforova et al., and other neoplasms as reported by The Cancer Genome Atlas fusion gene database. HTT hyalinizing trabecular tumor, ACC adrenocortical carcinoma, BLCA bladder urothelial carcinoma, BRCA breast invasive carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL cholangiocarcinoma, COAD colon adenocarcinoma, DLBC lymphoid neoplasm diffuse large B-cell lymphoma, ESCA esophageal carcinoma, GBM glioblastoma multiforme, HNSC head and neck squamous cell carcinoma, KICH kidney chromophobe, KIRC kidney renal clear cell carcinoma, KIRP kidney renal papillary cell carcinoma, LAML acute myeloid leukemia, LGG brain lower grade glioma, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, MESO mesothelioma, OV ovarian serous cystadenocarcinoma, PAAD pancreatic adenocarcinoma, PCPG pheochromocytoma and paraganglioma, PRAD prostate adenocarcinoma, READ rectum adenocarcinoma, SARC sarcoma, SKCM skin cutaneous melanoma, STAD stomach adenocarcinoma, TGCT testicular germ cell tumors, THCA thyroid carcinoma, THYM thymoma, UCEC uterine corpus endometrial carcinoma, UCS uterine carcinosarcoma, UVM uveal melanoma
Co-occurrence of a papillary thyroid carcinoma and a hyalinizing trabecular tumor in a single patient. The scanned histological section contains a small papillary carcinoma (2 mm, dashed circle line) and a hyalinizing trabecular tumor (main lesion, dashed semicircular line). Fluorescence in situ hybridization was performed and selected fields were scanned within the two lesions, thus demonstrating PAX8–GLIS3 fusions in the hyalinizing trabecular tumor (a), whereas no rearrangements were appreciated in the papillary carcinoma (b)
Of note, none of the eight cases subjected to RNA-sequencing harbored RET/PTC rearrangements.
Lack of PAX8–GLIS3 fusions in thyroid neoplasms
To determine whether PAX8–GLIS3 rearrangements would constitute a pathognomonic genetic alteration in the hyalinizing trabecular tumors, fluorescence in situ hybridization and RT-PCR were performed in the series of 237 control cases, which included the whole spectrum of conventional thyroid neoplasms and also encompassed a subgroup of 15 tumors of follicular derivation characterized by a prominent trabecular growth and hyalinization that was reminiscent of a hyalinizing trabecular tumor on sole morphological ground. None of these controls and potential differential diagnoses of hyalinizing trabecular tumor harbored the PAX8–GLIS3 rearrangement (Fig. 4e, Supplementary Table S3). In addition, none of the 9950 tumors from 33 cancer types from The Cancer Genome Atlas [33] were found to harbor the specific PAX8–GLIS3 fusion (Fig. 4f). These data provide evidence to suggest that the PAX8–GLIS3 fusion is pathognomonic for hyalinizing trabecular tumors.
Discussion
Here we present a validation of the recently reported PAX8–GLIS3 rearrangement [10] as a hallmark genomic feature of the rare hyalinizing trabecular tumor of the thyroid. In agreement with Nikiforova et al. [10] findings, all of the 34 histologically proven hyalinizing trabecular tumors of the present study harbored the PAX8–GLIS3 fusion. This gene rearrangement has been consistently demonstrated by different techniques, using specifically designed probes, which confirmed the occurrence of such rearrangement. In parallel, this fusion gene was not present in control thyroid tumors of various histologies, including an interesting series of trabecular tumors of follicular derivation that was specifically selected from our files for the purpose of testing the power of this novel fusion, as a potential differential diagnostic marker.
The origin of the hyalinizing trabecular tumor is unknown. No risk factors have been reported and the claimed relationship with papillary thyroid carcinoma had been challenged by the absence of BRAF mutations [6], and now definitely excluded by the specific PAX8–GLIS3 rearrangement (or PAX8–GLIS1, as reported by Nikiforova et al. [10]), that never occurred in the 300 papillary cancers so far analyzed in published and in the present series, with one exception in the Nikiforova’s series and in the The Cancer Genome Atlas series of 484 papillary carcinomas [35]. The case regarded a trabecular tumor focally reminiscent of a hyalinizing trabecular tumor that was eventually diagnosed as a papillary carcinoma and probably represents the same case used in the two different publications [10]. With regard to follicular adenoma and carcinoma, the similarities with hyalinizing trabecular tumor are even more scant on both morphological and genetic grounds. Besides RAS gene mutations, follicular adenomas and carcinomas were found to harbor a rearrangement of PAX8–PPARG genes that was originally reported to be restricted to malignant conditions, only [9], but subsequently identified also in a fraction of follicular adenomas, although never in hyalinizing trabecular tumors. Interestingly, the novel hyalinizing trabecular tumor-specific fusion is also correlated to the PAX8 gene, although the fusion occurred with another locus, mapping to chromosome 9p24.2 to produce the PAX8–GLIS3 rearrangement, or to chromosome 1 in the case of the rarer GLIS1 type fusion [10].
Although the newly discovered genetic alteration may not bear any impact in the therapeutic strategy of the patients, since surgery is generally sufficient to cure the tumor, the novel fusion product represents a relevant diagnostic tool of this rare tumor in surgical samples and, in particular, in preoperative cytology specimens, in which the diagnosis of hyalinizing trabecular tumor is reached in no more than 8% of cases [36]. In fact, the differential diagnosis from solid/trabecular variants of papillary carcinoma may occasionally pose problems due to the overlapping nuclear features of the two tumors, while genetically the two entities seem unrelated with a predominance of BRAF and RAS gene mutations in papillary cancer, but not in hyalinizing trabecular tumors [6]. In addition, even follicular neoplasms may resemble a hyalinizing trabecular tumor when the growth pattern is predominantly trabecular or solid, as seen in embryonal/fetal type follicular adenomas or in poorly differentiated carcinomas. The commonly shared thyroglobulin production is of no help in this setting of differential diagnosis, and the only useful hyalinizing trabecular tumor marker is the peculiar membrane pattern (artefactually) produced by Ki67 immunostaining, provided specific technical conditions are met (i.e., use of MIB1 monoclonal and room temperature incubation), in order to obtain the expected cell membrane reactivity [37].
Although diagnostic criteria for both hyalinizing trabecular tumors and its mimickers are apparently clear-cut, it is the experience of some of us that in the daily consultation practice, a certain degree of confusion exists among the different entities. The major source of controversies is the presence of more or less hyalinized stroma in an otherwise trabecular or solid/compact tumor cell proliferation of follicular derivation. Such a pattern of growth is shared by several primary tumors of various nature, including not only by follicular tumors (embryonal adenoma, solid variant of papillary carcinoma, Hurthle cell neoplasms, poorly differentiated carcinoma), but also by medullary carcinoma and intrathyroid parathyroid tumors. Apart from the differences in cell type and, obviously, in the ultimate immunoprofile of tumor cells, the trabecular structure is crucial for a diagnosis of a hyalinizing trabecular tumor: thick cords of elongated cells, arranged perpendicular to the major axis of the trabecula itself, contain variable amounts of hyaline collagenized material inside the trabeculae, to surround individual tumor cells. The general impression is that of a paraganglioma rather than a trabecular follicular-derived tumor, but the extent of hyalinization may change the overall picture and cause misdiagnoses. Indeed, all 15 trabecular cases, including four originally submitted as hyalinizing trabecular tumors, that we deliberately selected among the control cases lacked the PAX8–GLIS3 fusion. This supports the notion that the spectrum of trabecular thyroid neoplasms is relatively wide and heterogeneous, including a variety of growth patterns in which thick trabeculae merge with hyaline material depicting in the end more than one specific follicular derived, benign or malignant, entity or subtype. The novel described PAX8–GLIS3 fusion is a valuable hint to take the hyalinizing trabecular tumor apart from its mimickers.
Interestingly, four hyalinizing trabecular tumors cases had associated small papillary carcinomas, which did not harbor the GLIS3 fusion product. In addition, in the Nikiforova’s study [10] a link with upregulation of genes implicated in collagen type IV deposition was suggested, as the result of the gene rearrangement involving chromosomes 2 and 9, although the exact mechanisms remain to be elucidated. The abundance of basal membrane type material included collagen IV had been reported in prior immunohistochemical studies by Li and Rosai [38] and Papotti et al. [12]. These molecular and morphological findings may be of further help in taking apart real hyalinizing trabecular tumors from all the mimickers discussed above, care being taken to identify a predominant intratrabecular collagen deposition (rather than in the neoplastic stroma), since such a material seems to be the direct result of collagen-related gene upregulation in the fusion-positive neoplastic cells.
Finally, as demonstrated by Nikiforova et al. [10, 39] using ThyroSeq v3 gene panel in over 10,000 FNABs, this novel fusion may represent a relevant diagnostic marker of hyalinizing trabecular tumor in preoperative fine needle aspiration biopsies, especially to take it apart from other solid/trabecular thyroid tumors with prominent hyalinization. A correct preoperative characterization of hyalinizing trabecular tumors can result from molecular methods or from immunohistochemistry for GLIS3, to refrain from unnecessary surgery of indeterminate cytology trabecular/follicular nodules harboring the PAX8–GLIS3 fusion. Indeed, a proper diagnosis of hyalinizing trabecular tumor especially with respect to papillary thyroid carcinoma has important clinical implications, which are heavily related to the type of surgical strategy (since radical resection may not be necessary in hyalinizing trabecular tumor), as well as with the type of follow up or subsequent treatment (as no radiometabolic treatment is required in case of hyalinizing trabecular tumor).
In conclusion, the hyalinizing trabecular tumor represents a unique tumor based on its morphological (intratrabecular hyalinization, papillary cancer-like nuclei), immunohistochemical (Ki67/MIB1 expression at the cell membrane) and now also molecular (PAX8–GLIS3 fusion) features. The latter support the notion of separately classifying this tumor probably representing a peculiar variety of GLIS-rearranged hyalinizing trabecular adenoma. This is consistent with the original proposal by Carney et al. [11], who separated this tumor from both papillary carcinomas and the other conventional follicular thyroid neoplasms.
References
Jeong KJ, Park SY, Cho KH, Sohn JS, Lee J, Kim YK, et al. The Rho/ROCK pathway for lysophosphatidic acid-induced proteolytic enzyme expression and ovarian cancer cell invasion. Oncogene. 2012;31:4279–89.
Carney JA, Hirokawa M, Lloyd RV, Papotti M, Sebo TJ. Hyalinizing trabecular tumors of the thyroid gland are almost all benign. Am J Surg Pathol. 2008;32:1877–89.
Gowrishankar S, Pai SA, Carney JA. Hyalinizing trabecular carcinoma of the thyroid gland. Histopathology. 2008;52:529–31.
Cheung CC, Boerner SL, MacMillan CM, Ramyar L, Asa SL. Hyalinizing trabecular tumor of the thyroid: a variant of papillary carcinoma proved by molecular genetics. Am J Surg Pathol. 2000;24:1622–6.
Papotti M, Volante M, Giuliano A, Fassina A, Fusco A, Bussolati G, et al. RET/PTC activation in hyalinizing trabecular tumors of the thyroid. Am J Surg Pathol. 2000;24:1615–21.
Salvatore G, Chiappetta G, Nikiforov YE, Decaussin-Petrucci M, Fusco A, Carney JA, et al. Molecular profile of hyalinizing trabecular tumours of the thyroid: high prevalence of RET/PTC rearrangements and absence of B-raf and N-ras point mutations. Eur J Cancer. 2005;41:816–21.
Sheu SY, Vogel E, Worm K, Grabellus F, Schwertheim S, Schmid KW. Hyalinizing trabecular tumour of the thyroid-differential expression of distinct miRNAs compared with papillary thyroid carcinoma. Histopathology. 2010;56:632–40.
Acquaviva G, Visani M, Repaci A, Rhoden KJ, de Biase D, Pession A, et al. Molecular pathology of thyroid tumours of follicular cells: a review of genetic alterations and their clinicopathological relevance. Histopathology. 2018;72:6–31.
Kroll TG, Sarraf P, Pecciarini L, Chen CJ, Mueller E, Spiegelman BM, et al. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma [corrected]. Science. 2000;289:1357–60.
Nikiforova MN, Nikitski AV, Panebianco F, Kaya C, Yip L, Williams M, et al. GLIS rearrangement is a genomic hallmark of hyalinizing trabecular tumor of the thyroid gland. Thyroid. 2019;29:161–73.
Carney JA, Ryan J, Goellner JR. Hyalinizing trabecular adenoma of the thyroid gland. Am J Surg Pathol. 1987;11:583–91.
Papotti M, Riella P, Montemurro F, Pietribiasi F, Bussolati G. Immunophenotypic heterogeneity of hyalinizing trabecular tumours of the thyroid. Histopathology. 1997;31:525–33.
Marchio C, Geyer FC, Ng CK, Piscuoglio S, De Filippo MR, Cupo M, et al. The genetic landscape of breast carcinomas with neuroendocrine differentiation. J Pathol. 2017;241:405–19.
Guerini-Rocco E, Hodi Z, Piscuoglio S, Ng CK, Rakha EA, Schultheis AM, et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015;237:166–78.
Pareja F, Lee JY, Brown DN, Piscuoglio S, Gularte-Merida R, Selenica P, et al. The genomic landscape of mucinous breast cancer. J Natl Cancer Inst. 2019;111:djy216.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Geyer FC, Li A, Papanastasiou AD, Smith A, Selenica P, Burke KA, et al. Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas. Nat Commun. 2018;9:1816.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9.
Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76.
Narzisi G, Corvelo A, Arora K, Bergmann EA, Shah M, Musunuri R, et al. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun Biol. 2018;1:20.
Narzisi G, O’Rawe JA, Iossifov I, Fang H, Lee YH, Wang Z, et al. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods. 2014;11:1033–6.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131.
Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21.
Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 2018;8:174–83.
Zhang J, White NM, Schmidt HK, Fulton RS, Tomlinson C, Warren WC, et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 2016;26:108–18.
McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS. 2011;7:e1001138.
Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6.
Piscuoglio S, Burke KA, Ng CK, Papanastasiou AD, Geyer FC, Macedo GS, et al. Uterine adenosarcomas are mesenchymal neoplasms. J Pathol. 2016;238:381–8.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Shugay M, Ortiz de Mendibil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29:2539–46.
Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018;46:D1144–9.
Martelotto LG, De Filippo MR, Ng CK, Natrajan R, Fuhrmann L, Cyrta J, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–89.
Cancer Genome Atlas Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–90.
Saglietti C, Piana S, La Rosa S, Bongiovanni M. Hyalinizing trabecular tumour of the thyroid: fine-needle aspiration cytological diagnosis and correlation with histology. J Clin Pathol. 2017;70:641–7.
Leonardo E, Volante M, Barbareschi M, Cavazza A, Dei Tos AP, Bussolati G, et al. Cell membrane reactivity of MIB-1 antibody to Ki67 in human tumors: fact or artifact? Appl Immunohistochem Mol Morphol. 2007;15:220–3.
Li M, Carcangiu ML, Rosai J. Abnormal intracellular and extracellular distribution of basement membrane material in papillary carcinoma and hyalinizing trabecular tumors of the thyroid: implication for deregulation of secretory pathways. Hum Pathol. 1997;28:1366–72.
Nikiforova MN, Mercurio S, Wald AI, Barbi de Moura M, Callenberg K, Santana-Santos L, et al. Analytical performance of the ThyroSeq v3 genomic classifier for cancer diagnosis in thyroid nodules. Cancer. 2018;124:1682–90.
Acknowledgements
This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC, Milan, no. IG 20100 to MP), the Italian Ministry of Education, University and Research (MIUR ex-60%-2017, 2015HAJH8E and FPRC 5xmille 2015 MIUR-“Progetto Futuro” to CM). LA is funded by Fondazione Umberto Veronesi. We also acknowledge the funding by MIUR project “Dipartimenti di Eccellenza 2018–2022” (Project no. D15D18000410001) to the Department of Medical Sciences of the University of Turin, Italy.
JSR-F is funded by Breast Cancer Research Foundation. Research reported in this publication was funded in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant no. P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The funders of this study had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
CM has received personal/consultancy fees from Axiom Healthcare Strategies, Daiichi-Sankyo, and Bayer, outside the scope of the submitted work. JSR-F reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Roche and Genentech, outside the scope of the submitted work. All remaining authors have no conflicts of interest to declare.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Marchiò, C., Da Cruz Paula, A., Gularte-Merida, R. et al. PAX8–GLIS3 gene fusion is a pathognomonic genetic alteration of hyalinizing trabecular tumors of the thyroid. Mod Pathol 32, 1734–1743 (2019). https://doi.org/10.1038/s41379-019-0313-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0313-x
This article is cited by
-
Non-hyalinizing trabecular thyroid adenoma: a novel thyroid tumor with diagnostic pitfalls of hyalinizing trabecular adenoma and medullary thyroid carcinoma
Diagnostic Pathology (2023)
-
Overview of the 2022 WHO Classification of Thyroid Neoplasms
Endocrine Pathology (2022)