Abstract
Nasopharyngeal carcinoma (NPC), which is marked by a distinct distribution, is a common subtype of epithelial carcinoma arising from the nasopharyngeal mucosal lining. SRGN acts as an important and poor prognostic factor of NPC through multiple different mechanisms. However, the biological role and mechanism of SRGN in NPC remain unknown. Expression levels of miR-148a-5p, CREB1, FoxO1, and SRGN in NPC tissues and cell lines were tested by qRT-PCR or/and Western blot. The impacts of miR-148a-5p, CREB1, FoxO1, and SRGN on NPC cell viability, proliferation, migration, and invasion were estimated in vitro by CCK-8, colony formation, wound healing and Transwell experiments, and in vivo by a xenograft tumor model. JASPAR analysis was used to predict the binding activity of Foxo1 (CREB1) with the miR-148a-5p (SRGN) promoter, and the interaction was validated by EMSA and ChIP assays. The miR-148a-5p-CREB1 interaction was validated by a dual-luciferase reporter and RIP assays. CREB1 and SRGN were increased while miR-148a-5p was decreased in NPC. Silencing of SRGN and CREB1, as well as miR-148a-5p overexpression, repressed NPC tumor progression in vitro and in vivo. CREB1 promoted SRGN expression in NPC by targeting the promoter area of SRGN. Silencing of FoxO1 facilitated NPC tumor progression, while silencing of STAT3 repressed NPC tumor progression. FoxO1 bound to and regulated miR-148a-5p in NPC, and miR-148a-5p targeted CREB1. Additionally, FoxO1 knockdown abolished the downregulation of CREB1 and SRGN induced by STAT3 silencing. Our results suggest that STAT3 regulates SRGN and promotes the growth and metastasis of NPC through the FoxO1-miR-148a-5p-CREB1 axis.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC), originating from the nasopharyngeal mucosal lining, is one of the most common squamous cell carcinomas with a high probability of local invasion and distant metastasis1. Therefore, most NPC patients tend to exhibit regional lymph node or distant metastasis at diagnosis. NPC incidence shows a distinct geographical pattern in the world; it is one of the most frequently diagnosed malignancies in southern China, with an estimated incidence of 30 cases per 100,000 population, while only 1 case per 100,000 population is reported in the United States and Europe2,3. The main histological type of NPC is non-keratinizing carcinoma characterized by rich lymphocytic infiltration and Epstein-Barr virus (EBV) infection4. EBV DNA or clonal EBV genome can be invariably detected in NPC cells5. Moreover, certain prevalent EBV strains have been reported to be associated with an elevated risk of NPC6. These findings suggested a link between EBV infection and NPC tumorigenesis. Although NPC patients are usually sensitive to radiotherapy, their prognosis remains relatively poor due to the high probability of recurrence and distant metastasis7,8. Nevertheless, the mechanisms underlying NPC tumorigenesis are still not completely explored to date, and further study of these mechanisms is urgently needed.
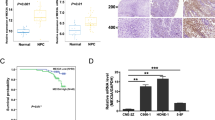
SRGN, predominately present in endothelial cells, cancer cells, and embryonic stem cells, is a low molecular weight glycoprotein linked to the storage and release of multiple proteases, cytokines and chemokines9,10. The highly expressed SRGN was found in metastatic NPC cells, knockdown of SRGN inhibited NPC cell metastasis while overexpression of SRGN promoted it, suggesting SRGN might be a promising therapeutic target for NPC11,12.
Signal transducer and activator of transcription 3 (STAT3) is an important transcription factor that belongs to the STAT protein family13. EBV infection can cause the activation of STAT3, and thus, facilitating the progression of NPC14,15,16. However, the underlying mechanisms of this effect remain largely unknown. FoxO1 is a critical member of the FOXO subfamily of the Forkhead/winged helix superfamily and is a transcription factor in mammalian cells17. It has been reported to act as a tumor repressor of NPC on the basis of its effects on modulating cell survival, metabolism and differentiation18. Considering the role of STAT3 and FoxO1 in NPC, and the interaction between STAT3 and FoxO1 was proved to be an important mechanism in the progression of various of cancers19,20, we believed the axis of STAT3/FoxO1 axis might also play a role in NPC. We also want to know if SRGN is involved in the function of STAT3/FoxO1 axis in NPC.
MicroRNAs (miRNAs) are key members of the noncoding RNA family, and accumulating evidence has shown that they regulate NPC tumor growth and metastasis both in vitro and in vivo21,22. MiR-148a-5p acts as an antitumor agent in breast cancer and gastric cancer23,24. Moreover, FoxO1 was demonstrated to be a transcription factor for miR-148a transactivation25. Additionally, we found that CREB1 is a target gene of miR-148a-5p by using TargetScan prediction and that CREB1 targets the promoter region of SRGN through JASPAR analysis. Therefore, we examined the relationship between STAT3, FoxO1, miR-148a-5p, CREB1 and SRGN in NPC in the present study.
Materials and methods
NPC tissues and cell lines
Thirty NPC tissues and thirty healthy control tissues were diagnosed at the People’s Hospital of Guangxi Zhuang Autonomous Region from 2017 to 2019 and stored at −80 °C. This research was approved by the Ethics Committee of the People’s Hospital of Guangxi Zhuang Autonomous Region, and informed consent was supplied by every subject involved in this study (No. 2018-47). NPC cell lines (CNE2, HNE1, and HONE1) were obtained from the Institute of Cancer, Central South University. The NPC cell line C666-1 and the normal immortalized nasopharyngeal epithelial cell line (NP69) were provided by Professor Sai-Wah Tsao from University of Hong Kong. SUNE-1 and 5-8F cells were kindly provided by Professor Musheng Zeng from Sun Yat-Sen University. All cell types were cultured in modified DMEM (10% fetal calf serum and 1% ampicillin/streptomycin) and maintained in an incubator with 95% O2 and 5% CO2 at 37 °C.
Quantitative real-time PCR (RT-PCR) analysis
Total RNA was extracted from NPC tissues and cells using TRIzol reagent (Invitrogen, USA) following the manufacturer’s instructions. After examination of its quality and concentration, the isolated RNA was used to generate cDNA using a Gibco BRL kit (Life Technologies, USA). qRT-PCR was conducted following standard protocols of the SYBR-GreenPCR kit (Takara, Japan) on an ABI7500 Fast Real-Time PCR System (PE Applied Biosystems). Sequences of primers are shown in Table 1.
Western blot
Total proteins of NPC tissues and cells were separated using high-speed centrifugation at 12,000 × g/min for 15 min after incubation in RIPA buffer (Beyotime, China). Protein samples (40 μg) were separated by 10% SDS-PAGE. Targeted proteins were transferred to nitrocellulose membranes followed by a 2-h incubation with 5% low-fat milk. Afterwards, membranes were incubated with 5% low-fat milk containing the indicated primary antibodies against CREB1 (rabbit, 1:500, ab32515, Abcam), SRGN (rabbit, 1:2000, ab156991, Abcam), FoxO1 (rabbit, 1:4000, ab10382, Abcam), STAT3 (rabbit, 1:5000, ab119352, Abcam), and β-actin (rabbit, 1:5000, ab179467, Abcam). After 8 h of incubation with primary antibodies, the membranes were probed with the indicated secondary antibodies for another 2 h. Signals were visualized using an enhanced chemiluminescence reagent (EMD Millipore, USA).
Establishment of stable NPC cell lines
Specific shRNAs against SRGN (shSRGN-1 and shSRGN-2), CREB1 (shCREB1-1 and shCREB1-2), FoxO1 (shFoxO-1 and FoxO1-2), and STAT3 (shSTAT3-1 and shSTAT3-2) as well as their negative control (shNC) were all supplied by Sigma. MiR-148a-5p mimics and negative control (NC) mimics were also obtained from GenePharma. HONE1 and 5-8F cells were cultured in DMEM in 24-well plates for 8 h followed by transfection with a specific lentiviral vector using the BLOCK-iT Lentiviral Pol II miR RNAi system (Invitrogen, USA). After three weeks of transfection, a single colony was selected and identified. After infection with lentivirus, cells were subjected for selection using 3 μg/ml puromycin for 24 h to remove any remaining non-infected cells. After screen, cells were collected for further experiments.
Evaluation of cell proliferation
The proliferation of treated NPC cells was evaluated through the Cell Counting Kit-8 (CCK-8, Solarbio, Beijing, China) and colony formation experiments. CCK-8-treated NPC cells (2 × 105 cells/well) were seeded into 96-well plates containing FBS and DMEM. Next, 10 μl of CCK-8 solution was added to every well. After 2 h of incubation at 37 °C, the absorbance of every single well was determined at 490 nm. For colony formation, treated NPC cells at the exponential growth phase were harvested and seeded in 35-mm petri dishes followed by two weeks of culture. Next, the visualized colonies were stained with crystal violet for 10 min after fixation in methanol. The number of colonies was manually counted.
Evaluation of cell migration
A wound healing assay was conducted on treated HONE1 and 5-8F cells to evaluate cell migration. In brief, cells were collected after treatment and then plated in 35-mm petri dishes. Treated HONE1 and 5-8F cells were cultured at 37 °C until 100% confluence and then received a straight scratch on the surface of the NPC cell layer. Images were photographed after 0 and 24 h of scratching.
Evaluation of cell invasion
Transwell chambers with an 8-μm pore size (Corning Incorporated, USA) coated with Matrigel were used to estimate the invasion of HONE1 and 5-8F cells. Briefly, 500 μl of serum-free DMEM containing 2 × 105 treated HONE1 and 5-8F cells was added to the upper chamber, while 500 μl of DMEM including serum (10%) was added to the lower chamber. After 24 h of culture, cells remaining on the upper surface were discarded, and those cells on the bottom surface were fixed and stained with 5% crystal violet followed by manual number counting.
Xenograft tumor growth and in vivo metastasis evaluation
Eight-week-old BALB/c nude mice (male) were supplied by Hunan Slake Jingda Laboratory Animal Co., Ltd., and animal manipulation was approved by the ethics committee of our institute (No. 2018-47). Animals were maintained in a pathogen-free environment with free access to water and food. HONE1 and 5-8F cells transfected with the indicated oligonucleotides, including shSRGNs, shCREB1s, shFoxO1s, shSTAT3s, and miR-148a-5p mimics, were subcutaneously delivered into the left flank of the mice. The tumor volume was measured every 10 days and then weighed 40 days after inoculation. For in vivo metastasis evaluation, 2 × 106 treated NPC cells were injected into nude mice via the tail vein. These treated animals were sacrificed 40 days after inoculation, and liver and lung tissues were collected for metastatic nodule analysis. After fixation with 4% paraformaldehyde overnight, liver and lung tissues were cut into 5-μm slices, and histopathologic analysis was conducted using haematoxylin and eosin staining.
Validation of protein and promoter binding
The binding activity of CREB1 on the promoter region of SRGN was estimated by electrophoretic mobility shift assay (EMSA) using an EMSA kit (BersinBio, China) according to the manufacturer’s instructions in a reaction system containing nuclear extracts and specific biotin probes. Moreover, ChIP was performed to further verify the interaction between CREB1 or FoxO1 and the promoter region of SRGN or miR-148a-5p using a ChIP assay kit (Thermo Scientific, USA). DNA fragments of treated NPC cells were isolated and digested with micrococcal nuclease. Anti-CREB1 or IgG was added to the reaction system for immunoprecipitation. Subsequently, qRT-PCR was used to analyze the enrichment after elution and purification.
Co-immunoprecipitation (Co-IP) assay
The interplay between FoxO1 and Runx2 was verified by Co-IP in 293T cells. In brief, 293T cells were transfected with Myc-tagged FoxO1 (Myc-FoxO1) and Flag-tagged Runx2 (Flag-Runx2). After 48 h of transfection, cells were harvested and washed with precooled phosphate buffer solution followed by lysis in extraction buffer (pH 8.0, 50 mM Tri-HCl, 1 mM EDTA, 150 mM NaCl and protease repressors). Cell lysates were then incubated overnight with monoclonal anti-Flag and anti-Myc antibodies at 4 °C, and SDS-PAGE was performed to isolate immunoprecipitated samples followed by transfer to PVDF membranes (Millipore, USA). After blocking with 5% non-fat milk for 2 h, the membranes were probed with horseradish peroxidase-conjugated secondary antibodies against Flag and Myc. Bands were visualized using chemiluminescence (ECL) reagent.
Dual-luciferase reporter assay
The interaction between the 3’UTR of CREB1 and miR-148a-5p was tested by dual-luciferase reporter assays. Briefly, two wild-type (WT) putative miR-148a-5p binding sites (BS1 and BS2) of the CREB1 3’UTR were amplified and inserted into the pGL3 vector to establish recombinant luciferase reporter plasmids named CREB1-WT. The matched mutant (Mut) miR-148a-5p binding sites were also cloned into the pGL3 vector to establish mutant recombinant luciferase reporter plasmids named CREB1-Mut1 (BS1 mutation), CREB1-Mut2 (BS2 mutation) and CREB1-Mut1&2 (both the BS1 and BS2 mutations). To examine the interaction between miR-148a-5p and the CREB1 3’UTR, HONE1 and 5-8F cells were co-transfected with CREB1-WT, CREB1-Mut1, CREB1-Mut2, CREB1-Mut1&2 and miR-148a-5p mimics or NC mimics. After 24 h of co-transfection, the luciferase activity of HONE1 and 5-8F cells was determined by the Dual-Luciferase Reporter Assay System (Promega, USA).
RNA immunoprecipitation (RIP)
RIP was used to further examine the interaction between CREB1 and miR-148a-5p. In brief, HONE1 and 5-8F cells were transfected with MS2bs-Rluc, MS2bs-CREB1-3’UTR and MS2bs-CREB1-3’UTR-Mt. After 48 h of transfection, the RIP assay was carried out using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). MiR-148a-5p was quantified using qRT-PCR after the RNA complexes were purified
Statistical analysis
Data are shown as the mean ± standard deviation (SD). Statistical analysis was completed in GraphPad Prism (Version 7.0, USA). Two groups were compared with Student’s t test, multiple experimental groups were compared using one-way analysis of variance (ANOVA) or two-way ANOVA, and p < 0.05 was considered statistically significant.
Results
CREB1 and SRGN showed high expression, while miR-148a-5p showed lower expression in NPC
We first tested the expression of miR-148a-5p, CREB1, SRGN, and FoxO1 in 30 NPC tissues and 30 healthy controls by qRT-PCR and found that miR-148a-5p and FoxO1 were lower while the mRNAs of CREB1 and SRGN were higher in NPC tissues than in normal nasopharyngeal mucosal tissues (Fig. 1A). In addition, we demonstrated that miR-148a-5p was downregulated in six NPC cell lines (CNE2, HNE1, 5-8F, HONE1, C666-1, and SUNE-1) compared to the NP69 cell line (Fig. 1B). The protein expression of CREB1 and SRGN was higher while FoxO1 was lower in NPC cell lines than in the NP69 cell line (Fig. 1C). Thus, these findings suggested that miR-148a-5p, CREB1 and SRGN were dysregulated in NPC.
A The relative expression levels of CREB1, SRGN, miR-148a-5p and FoxO1 in NPC tissues were analyzed by qRT-PCR, and the control group included healthy nasopharyngeal mucosal tissues, n = 30. B Relative expression levels of miR-148-5p in six NPC cell lines (CNE2, HNE1, 5-8F, HONE1, C666-1, and SUNE-1) and one normal immortalized nasopharyngeal epithelial cell line (NP69) were analyzed by qRT-PCR, n = 3. C Protein expression levels of CREB1, SRGN and FoxO1 in six NPC cell lines (CNE2, HNE1, 5-8F, HONE1, C666-1, and SUNE-1) and one normal immortalized nasopharyngeal epithelial cell line (NP69) were detected by western blotting. GAPDH was used as an internal control, n = 3. All the results are shown as the mean ± SD of three different experiments performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
SRGN silencing repressed NPC tumor progression
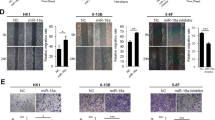
The role of SRGN in NPC tumor progression was further examined. Based on the elevated level of SRGN, we decided to exogenously silence its expression in the HONE1 and 5-8F cell lines. As shown in the results, SRGN protein was decreased by 3.5 times in HONE1 and by 1.8 times in 5-8F cell lines after transfection of shSRGN-1, moreover, it was decreased by 5 times in HONE1 and by 2.9 times in 5-8F cell lines after transfection of shSRGN-2 (Fig. 2A). The impact of SRGN silencing on NPC cell proliferation activity was estimated by the CCK-8 and colony formation experiments. The CCK-8 results indicated that transfection of shSRGN-1 reduced the viability of HONE1 and 5-8F cells to around 68% and 71%, respectively, shSRGN-2 transfection reduced the viability HONE1 and 5-8F cells to around 73% and 62%, respectively (Fig. 2B). The colony numbers of shSRGN-1-transfected HONE1 and 5-8F cells were reduced to around 40% and 36%, respectively, of shNC group, same trend was observed in shSRGN-2-transfected cell (Fig. 2C). The effects of SRGN silencing on NPC cell migration and invasion were subsequently evaluated by wound healing and Transwell assays. SRGN silencing resulted in a significant repression of HONE1 and 5-8F cell migration and invasion (Fig. 2D and E). Additionally, we found that the protein expression of CREB1 in sh-SRGN-1 transfected HONE1 and 5-8F cell was reduced to 49% and 55% of shNC-transfected cells, respectively, same trend was observed in shSRGN-2-transfected cell (Fig. 2F). A xenograft tumor assay was further performed to examine the role of SRGN silencing in tumor growth in vivo. shNC-, shSRGN-1- and shSRGN-2-transfected HONE1 and 5-8F cells were subcutaneously delivered into nude mice. The weight and volume of tumors formed by SRGN-silenced HONE1 and 5-8F cells were markedly decreased compared to those formed by shNC-transfected cells (Supplementary Fig. S1A–C). shNC-, shSRGN-1- and shSRGN-2-transfected 5-8F cells were then inoculated into nude mice via tail vein injection. After ten weeks of inoculation, lung and liver metastasis were apparently reduced in animals injected with SRGN-silenced cells compared to negative control cells (Supplementary Fig. S1D, E). Similar results were observed in histologic analysis (Supplementary Fig. S1F). These results indicated that SRGN silencing repressed NPC tumor progression.
A Knockdown efficiency of SRGN protein by shRNAs (shSRGN-1 and shSRGN-2) was examined by western blotting. B After 0, 1, 2, and 3 days of SRGN shRNA transfection, proliferation of HONE1 and 5-8F cells were examined by the CCK-8 assay. C The colony formation ability of HONE1 and 5-8F cells transfected with shNC and SRGN shRNAs was estimated. D Wound healing and E Transwell assays were conducted on shNC- and SRGN shRNA-transfected HONE1 and 5-8F cells to evaluate their migratory and invasive abilities, respectively. F Protein levels of CREB1 were measured by western blotting in sh-NC and SRGN shRNA-transfected HONE1 and 5-8F cells. All the results are shown as the mean ± SD (n = 3), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
CREB1 silencing repressed NPC tumor progression and decreased the expression of SRGN
Next, the same strategy was adopted to study the effects of CREB1 silencing on NPC tumor progression. Western blot analysis of CREB1 showed that transfection of CREB1 shRNAs (shCREB1-1 and shCREB1-2) resulted in a significant downregulation of CREB1 compared to shNC transfection (Fig. 3A). The growth of CREB1-silenced HONE1 and 5-8F cells was markedly repressed, as shown by the CCK-8 assay (Fig. 3B). The colony numbers of CREB1-silenced HONE1 and 5-8F cells were significantly reduced (Fig. 3C). In wound-healing and Transwell assays, we revealed that CREB1 knockdown caused a remarkable repression of the migratory and invasive abilities of HONE1 and 5-8F cells (Fig. 3D, E). Additionally, we demonstrated that SRGN protein was downregulated in the CREB1-silenced group compared to the shNC group (Fig. 3F). The NPC tumor growth repressive effects of CREB1 shRNAs were further validated by the xenograft tumor growth assay. After inoculation of CREB1-silenced HONE1and 5-8F cells into nude mice, we found that tumors formed by CREB1-silenced HONE1 and 5-8F cells were significantly smaller than those formed by shNC-treated cells (Supplementary Fig. S2A–C). Moreover, the metastatic nodules of the liver and lung observed in mice inoculated with CREB1-silenced 5-8F cells were significantly reduced compared to those in the shNC group (Supplementary Fig. S2D, E). Consistent results were observed in histopathologic analysis (Supplementary Fig. S2F). These findings indicated that CREB1 silencing repressed NPC tumor progression and decreased the expression of SRGN.
A The knockdown efficiency of CREB1 protein was tested via western blotting. B After 0, 1, 2, and 3 days of CREB1 shRNA transfection, a CCK-8 assay was performed in HONE1 and 5-8F cells to examine cell proliferation. C The colony formation ability of CREB1-silenced HONE1 and 5-8F cells was determined. D Wound healing and E Transwell assays were carried out to estimate the migratory and invasive capacities of HONE1 and 5-8F cells after CREB1 knockdown, respectively. F The protein level of SRGN was detected by western blotting in HONE1 and 5-8F cells transfected with sh-NC and CREB1 shRNAs. All the results are shown as the mean ± SD (n = 3), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
CREB1 regulated SRGN expression by targeting its promoter
By using western blot analysis, we found that CREB1 overexpression markedly increased the protein expression of CREB1 and SRGN (Fig. 4A). Moreover, JASPAR analysis predicted a putative CREB1 binding site in the promoter region of SRGN (BS: 1442-1454) (Fig. 4B). EMSA was then performed to validate the binding of the CREB1 protein and the SRGN promoter. As shown in the results, the CREB1 protein could form a DNA-protein complex with a native probe containing putative CREB1 binding sites (Fig. 4C). Additionally, ChIP experiments showed that the CREB1 binding site in the SRGN promoter area was functional (Fig. 4D). These results suggested that CREB1 regulated SRGN expression by binding to its promoter.
A Western blot analysis of CREB1 and SRGN in 5-8F cells transfected with NC and CREB1. B The targeting site of CREB1 at the promoter region of SRGN was predicted by JASPAR analysis. C EMSA was performed to validate the interplay between the CREB1 protein and SRGN promoter in 5-8F cells. D ChIP-PCR was employed to assess CREB1 binding to the SRGN promoter in CREB1-silenced 5-8F cells. All the results are shown as the mean ± SD (n = 3), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01.
miR-148a-5p overexpression repressed NPC tumor progression
Next, we investigated the biological functions of miR-148a-5p in NPC tumor progression by overexpressing it in NPC. The relative miR-148a-5p expression in HONE1 and 5-8F cells was significantly upregulated after miR-148a-5p mimics transfection compared to the NC mimics group (Fig. 5A). The growth of miR-148a-5p-overexpressing HONE1 and 5-8F cells was dramatically repressed, as shown by the CCK-8 and colony formation assays (Fig. 5B, C). Moreover, in wound healing and Transwell assays, miR-148a-5p overexpression significantly inhibited the migratory and invasive capacities of HONE1 and 5-8F cells (Fig. 5D, E). Through western blot analysis, we found that the protein expression levels of CREB1 and SRGN were sharply decreased in the miR-148a-5p-overexpressing group compared to the NC group (Fig. 5F). Xenograft tumor growth assays demonstrated that the tumors formed by miR-148a-5p-overexpressing HONE1 and 5-8F cells were dramatically smaller than those formed by NC mimics-transfected cells (Supplementary Fig. S3A–C). NC mimics- and miR-148a-5p mimics-transfected 5-8F cells were then inoculated into nude mice via tail vein injection. After ten weeks of inoculation, the metastatic nodules in the lung and liver were significantly reduced in the miR-148a-5p group compared to the NC mimics group (Supplementary Fig. S3D, E). Similar results were observed in the histopathologic analysis performed in liver and lung tissues of animals injected with miR-148a-5p mimics-overexpressing cells through the tail vein (Supplementary Fig. S3F). Overall, miR-148a-5p overexpression repressed NPC tumor progression.
A Overexpression efficiency of miR-148-5p in HONE1 and 5-8F cells was tested by qRT-PCR. B The proliferation of HONE1 and 5-8F cells was evaluated by the CCK-8 assay after 0, 1, 2, and 3 days of miR-148a-5p mimics transfection. C After transfection with mimics NC and miR-148a-5p mimics, a colony formation assay was performed in HONE1 and 5-8F cells to test the cell colony formation ability. D, E Wound healing and Transwell assays were carried out to estimate the migratory and invasive capacities of HONE1 and 5-8F cells after miR-148a-5p mimics transfection, respectively. F Protein levels of CREB1 and SRGN were examined by western blotting in HONE1 and 5-8F cells transfected with miR-148a-5p mimics. All the results are shown as the mean ± SD (n = 3), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
FoxO1 silencing facilitated NPC tumor progression
To investigate whether FoxO1 is involved in NPC tumorigenesis, we examined the effects of FoxO1 knockdown on NPC tumor progression in vitro and in vivo. The knockdown efficiency of FoxO1 shRNAs (shFoxO-1 and shFoxO-2) was tested by western blotting (Fig. 6A). In vitro experiments demonstrated that FoxO1 silencing markedly facilitated cell growth (Fig. 6B), colony formation (Fig. 6C), migration (Fig. 6D) and invasion (Fig. 6E). Additionally, through qRT-PCR and western blot analysis, we observed a significant downregulation of miR-148a-5p and a remarkable upregulation of CREB1 and SRGN in the FoxO1 shRNA-transfected groups (Fig. 6F, G). Unexpectedly, Runx2 shRNA transfection only decreased the expression of Runx2 in HONE1 and 5-8F cells without affecting the protein levels of CREB1 and SRGN (Fig. 6H). The results from the in vivo xenograft tumor growth assay further confirmed the effects of FoxO1 knockdown on promoting NPC tumor growth (Supplementary Fig. S4A–C). By inoculating shNC- and FoxO1 shRNA-transfected 5-8F cells into nude mice via tail vein injection, we demonstrated that FoxO1 knockdown increased the number of metastatic nodules in the lung and liver, indicating that FoxO1 knockdown promoted NPC metastasis in vivo (Supplementary Fig. S4D, E). This conclusion was further supported by histopathologic analysis (Supplementary Fig. S4F). These findings indicated that FoxO1 might act as a tumor repressor of NPC via the CREB1-SRGN axis.
A The knockdown efficiency of FoxO1 protein was verified by western blotting. B The proliferation of HONE1 and 5-8F cells was analyzed by the CCK-8 assay after 0, 1, 2, and 3 days of FoxO1 shRNA transfection. C The effects of FoxO1 knockdown on the colony formation ability of HONE1 and 5-8F cells were assessed by the colony formation assay. D Wound healing and E Transwell assays were used to analyze the migratory and invasive capacities of HONE1 and 5-8F cells after FoxO1 knockdown, respectively. F Protein expression of CREB1 and SRGN was detected in tumors formed by FoxO1-silenced HONE1 and 5-8F cells by western blotting. G Relative miR-148a-5p expression was evaluated by qRT-PCR in tumors formed by FoxO1-silenced HONE1 and 5-8F cells. All the results are shown as the mean ± SD (n = 4), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
FoxO1 bound to the promoter of MIR148, while miR-148a-5p targeted CREB1
Interestingly, we found two miR-148a-5p binding sites in the 3’-UTR of CREB1 based on the prediction results of starBase (Fig. 7A). Subsequently, a dual-luciferase reporter assay was employed to validate the interplay between miR-148a-5p and CREB1. We established luciferase reporter plasmids by inserting the two mutant miR-148a-5p binding sites into the pGL3 vector separately or together and named them CREB1-Mut1, CREB1-Mut2 and CREB1-Mut1&2. Transfection of miR-148a-5p mimics, but not NC mimics, dramatically reduced the luciferase intensity of cells driven by CREB1-WT, while no change was observed in cells co-transfected with miR-148a-5p mimics and CREB1-Mut1 or CREB1-Mut2 (Fig. 7B). To further validate the direct binding between miR-148a-5p and the CREB1 3’UTR, we performed a RIP assay. The results indicated that miR-148a-5p was predominately enriched in the MS2bs-CREB1 group (Fig. 7C). Additionally, we observed a significant upregulation of primary and mature forms of miR-148a-5p in FoxO1-overexpressing cells (Fig. 7D). JASPAR prediction showed three putative FoxO1 binding sites (BS1, BS2 and BS3) in the promoter region of MIR148 (miR-148a-5p corresponding gene) (Fig. 7E). Then, a ChIP assay was performed to verify the interaction between FoxO1 and the three putative binding sites of MIR148. As demonstrated in the results, FoxO1 was significantly enriched in the BS3 group, indicating that FoxO1 is only bound to BS3 (Fig. 7F). Then, the wild-type and mutant BS3 fragments were amplified and inserted into the pGL3 vector to establish the luciferase reporter plasmids pGL3-MIR148 WT and pGL3-MIR148 Mut. The luciferase activity driven by pGL3-MIR148 WT was dramatically attenuated by the transfection of shFoxO1 but not shNC (Fig. 7G). These findings indicated that FoxO1 binds to the promoter of MIR148, while miR-148a-5p targets CREB1.
A The binding sites between miR-148a-5p and CREB1 were predicted by starBase software. B A dual-luciferase reporter assay was conducted to validate the interplay between miR-148a-5p and CREB1 using two individual binding sites predicted by starBase software. C The RIP assay was performed to further confirm the interplay between miR-148a-5p and CREB1. D qRT-PCR was carried out to detect the expression of primary and mature forms of miR-148a-5p after FoxO1 overexpression. E The targeting site of FoxO1 at the promoter region of MIR148 was predicted by JASPAR analysis. F ChIP assays were conducted to verify the binding between FoxO1 and MIR148. G The interplay between MIR148 and FoxO1 was verified by a dual-luciferase reporter assay. All the results are shown as the mean ± SD (n = 3), and three different experiments were performed in triplicate. **P < 0.01, ***P < 0.001. ns no significant.
STAT3 silencing repressed NPC tumor progression and regulated CREB1 and SRGN through FoxO1
To explore the role of STAT3 during NPC tumorigenesis, we evaluated the effects of STAT3 knockdown on NPC tumor progression in vitro and in vivo. The knockdown efficiency of STAT3 shRNAs was validated by western blotting (Fig. 8A). The results from in vitro assays indicated that STAT3 silencing resulted in a significant suppression of NPC cell proliferation, colony formation, migration and invasion (Fig. 8B–E). Western blot assays showed that STAT3 silencing resulted in a significant upregulation of FoxO1 but caused a remarkable downregulation of CREB1 and SRGN (Fig. 8F). Additionally, we also found that STAT3 silencing caused a significant upregulation of miR-148a-5p by qRT-PCR (Fig. 8G). However, the dysregulation of miR-148a-5p, FoxO1, CREB1 and SRGN caused by STAT3 silencing could be rescued by FoxO1 knockdown (Fig. 8H, I). The in vivo suppressive effects of STAT3 shRNAs on NPC tumor growth were verified by a xenograft tumor growth assay (Supplementary Fig. S5A–C). Moreover, the in vivo repressive effects of STAT3 shRNAs on NPC metastasis were supported by the results from the tail vein injection assay and histopathologic analysis (Supplementary Fig. S5D–F). We conducted rescue experiments to rule out off target effects of CREB1 gene knockdown and confirm our model. STAT3 and miR-148a-5p mimics or shCREB1 were co-transfected into 5-8F and HONE1 cells, followed by functional assays. The overexpression efficiency of STAT3 in 5-8F and HONE1 cells was verified by western blotting (Fig. 9A). By using CCK-8 and colony formation assays, we found that miR-148a-5p overexpression and CREB1 knockdown weaken the function of promoting cell proliferation of STAT3 overexpression (Fig. 9B, C). By using wound healing and/or Transwell assays, we demonstrated that cell migration and invasion were significantly repressed by miR-148a-5p overexpression and CREB1 knockdown, moreover, overexpression of miR-148a-5p and knockdown of CREB1 weaken the function of promoting invasion and metastasis of overexpression of STAT3 (Fig. 10A, B). Additionally, we examined the expression of miR-148a-5p, STAT3, FoxO1, CREB1, and SRGN in 5-8F and HONE1 cells transfected with STAT3 and miR-148a-5p mimics or shCREB1. STAT3 overexpression caused a significant downregulation of miR-148a-5p and reversed the upregulation of miR-148a-5p induced by miR-148a-5p mimics transfection (Fig. 10C). Moreover, the significant downregulation of CREB1 and SRGN caused by overexpression of STAT3 can be blocked by overexpression of mir-148a-5p and CREB1 (Fig. 10D). These results indicated that STAT3 silencing repressed NPC tumor progression and regulated the expression of miR-148a-5p, CREB1 and SRGN through FoxO1.
A Knockdown efficiency of STAT3 protein by shRNAs was examined by western blotting. B, C CCK-8 and colony formation assays were utilized to detect cell proliferation after STAT3 knockdown. D, E Wound healing and Transwell assays were performed to analyze cell migration and invasion after STAT3 knockdown. F Protein levels of FoxO1, CREB1 and SRGN were detected by western blotting in STAT3-silenced cells. G Relative expression of miR-148a-5p was examined in cells transfected with shNC and shSTAT3. H Protein levels of STAT3, FoxO1, CREB1 and SRGN were detected by western blotting in STAT3- and/or FoxO1-silenced cells. I The relative expression of miR-148a-5p was examined in cells transfected with shSTAT3 and/or shFoxO1. All the results are shown as the mean ± SD (n = 3), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
A The overexpression efficiency of STAT3 in 5-8F and HONE1 cells was verified by western blotting. B MTT and C colony formation assays were performed to detect the proliferation of 5-8F and HONE1 cells transfected with STAT3 and miR-148a-5p mimics or shCREB1. All the results are shown as the mean ± SD (n = 4), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, *** P < 0.001.
A Wound healing and B Transwell assays were performed to measure the cell migration and invasion of 5-8 F and HONE1 cells transfected with STAT3 and miR-148a-5p mimics or shCREB1. C Relative expression of miR-148a-5p was examined by qRT-PCR in each group. D Protein expression of STAT3, FoxO1, CREB1, and SRGN was assessed by western blotting in each group. All the results are shown as the mean ± SD (n = 4), and three different experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
SRGN ovexpression abolished the repressive effects of miR-148a-5p mimic and shCREB1 on NPC cell proliferation, migration, and invasion
Rescue assays were performed to further confirm the regulation axis of miR-148a-5p/CREB1/SRGN in NPC cell. By transfecting HONE1 and 5-8F cells with SRGN overexpression plasmid, we found an 80% increase in SRGN expression level compared to NC group (Fig. 11A). Overexpression of SRGN in HONE1 and 5-8F cells increased cell proliferation viability, while co-transfection with SRGN and miR-148a-5p mimics or shCREB1 abolished the promotive effects of SRGN overexpression on NPC cell proliferation (Fig. 11B, C). In addition, SRGN overexpression was found to promoted the migration and invasion of HONE1 and 5-8F cells, while co-transfection with SRGN and miR-148a-5p mimics or shCREB1 abolished the promotive effects of SRGN overexpression on NPC cell migration and invasion (Fig. 11D, E). Co-transfection with SRGN, miR-148a-5p mimics, and shCREB1 further inhibited cell proliferation, migration, and invasion of HONE1 and 5-8F cells (Fig. 11B–E). These results further confirmed the regulation axis of miR-148a-5p/CREB1/SRGN in NPC cell.
A Western blot analysis of SRGN protein level in HONE1 and 5-8F cells transfected with SRGN overexpression plasmid. B CCK-8 and C colony formation assays were employed to evaluate NPC cell proliferation viability after transfection with SRGN, miR-148a-5p mimics, or/and shCREB1. D Wound-healing and E transwell assays were carried out to test the effects of transfection with SRGN, miR-148a-5p mimics, or/and shCREB1 on NPC cell migration and invasion, respectively.
Discussion
MiR-148a-5p has been reported to be associated with the aetiology of several kinds of human tumors23,24. Nevertheless, its role and mechanism in NPC tumorigenesis have not been investigated. In this study, we found that miR-148a-5p was expressed at low levels in NPC and acted as an NPC suppressor in vitro and in vivo. Furthermore, we demonstrated that FoxO1, miR-148a-5p and CREB1 function as an axis during NPC progression. Additionally, STAT3 was revealed to facilitate NPC tumor progression by regulating SRGN expression through the FoxO1/miR-148a-5p/CREB1 axis.
CAMP-response element-binding protein (CREB1) is a transcription factor involved in modulating gene transcription by phosphorylation and dephosphorylation. Dysregulation of CREB1 is frequently observed in numerous human cancers, including NPC26. However, the biological role of CREB1 in NPC remains largely unclear. Here, we found that CREB1 silencing inhibited NPC tumor progression. Mechanistically, miR-148a-5p was revealed to negatively regulate SRGN expression by targeting the CREB1 3’-UTR and inhibiting its expression.
Recently, SRGN was revealed to promote NPC metastasis by inducing an epithelial-mesenchymal transition12. In addition, SRGN increased the production and release of transforming growth factor-β2 by activating the CD44/CREB1 axis in breast cancer cells27. As an important cell adhesion molecule involved in tumor progression and metastasis, CD44 has been reported to interact with SRGN in non-small-cell lung cancer28. In this study, SRGN was identified as an oncogene of NPC in vitro and in vivo. Mechanistically, CREB1 is bound to the promoter of SRGN and positively regulated SRGN. Knockdown of SRGN caused a significant downregulation of CREB1 and CD44. Taken together, these findings suggested that CREB1, SRGN and CD44 might form a regulatory loop in NPC.
Although FoxO1 has been well documented as a tumor suppressor in many cancers29, little is known about its role in NPC progression. Recently, FoxO1 was demonstrated to repress NPC cell growth by inhibiting cell cycle transition mediated by the PI3K/AKT cascade18. Consistently, in our study, we demonstrated that FoxO1 served as an NPC repressor in vitro and in vivo. Mechanistically, FoxO1 was revealed to interact with numerous miRNAs in various human cancers30,31,32. By using JASPAR analysis, we found three FoxO1 binding sites in the promoter region of miR-148a-5p. We observed a remarkable upregulation of both primary and mature forms of miR-148a-5p, which suggested that FoxO1 acted as an upstream positive regulator of miR-148a-5p in NPC. Thus, FoxO1 might function as a repressor of NPC through the miR-148a-5p/CREB1/SRGN axis. STAT3 is considered to be an oncogene in NPC. Consistent with previous studies, we supported the oncogenic role of STAT3 in NPC by providing in vitro and in vivo evidence. In addition, the interaction between FoxO1 and STAT3 has been well characterized by numerous studies in various human cancers19,20,33. Here, we demonstrated that FoxO1 silencing could abolish the downregulation of CREB1 and SRGN induced by STAT3 knockdown.
Taken together, these results provide strong evidence that STAT3 promotes NPC tumor progression by regulating the expression of SRGN through the FoxO1/miR-148a-5p/CREB1 axis (Fig. 12). Our findings reveal a potential therapeutic axis, which might contribute to the development of efficient drugs for NPC patients.
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Lu, J. et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 71, 225–233 (2011).
Cao, S. M., Simons, M. J. & Qian, C. N. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin. J. Cancer 30, 114–119 (2011).
Yu, M. C. & Yuan, J. M. Epidemiology of nasopharyngeal carcinoma. Semin. Cancer Biol. 12, 421–429 (2002).
Hau, P. M. et al. Targeting Epstein-Barr virus in nasopharyngeal carcinoma. Front. Oncol. 10, 600 (2020).
Tsao, S. W., Tsang, C. M. & Lo, K. W. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160270 (2017).
Hui, K. F. et al. High risk Epstein-Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int. J. Cancer 144, 3031–3042 (2019).
Mao, Y. P. et al. Prognostic factors and failure patterns in non-metastatic nasopharyngeal carcinoma after intensity-modulated radiotherapy. Chin. J. Cancer 35, 103 (2016).
Shen, L. J. et al. Subdivision of M category for nasopharyngeal carcinoma with synchronous metastasis: time to expand the M categorization system. Chin. J. Cancer 34, 450–458 (2015).
Kolseth, I. B. M. et al. Serglycin is part of the secretory repertoire of LPS‐activated monocytes. Immun. Inflamm. Dis. 3, 23–31 (2015).
Kolset, S. O. & Pejler, G. Serglycin: a structural and functional chameleon with wide impact on immune cells. J. Immunol. 187, 4927–4933 (2011).
Chia, C. S. et al. Serglycin expression: an independent marker of distant metastases in nasopharyngeal carcinoma. Head Neck 38, 21–28 (2016).
Li, X. J. et al. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 71, 3162–3172 (2011).
Fathi, N., Rashidi, G., Khodadadi, A., Shahi, S. & Sharifi, S. STAT3 and apoptosis challenges in cancer. Int. J. Biol. Macromol. 117, 993–1001 (2018).
Liu, J. et al. The effects of microRNA-98 inhibits cell proliferation and invasion by targeting STAT3 in nasopharyngeal carcinoma. Biomed. Pharmacother. 93, 869–878 (2017).
Lin, C. H., Chiang, M. C. & Chen, Y. J. STAT3 mediates resistance to anoikis and promotes invasiveness of nasopharyngeal cancer cells. Int. J. Mol. Med. 40, 1549–1556 (2017).
Gao, J. et al. Targeted regulationof STAT3 by miR-29a in mediating Taxol resistance of nasopharyngeal carcinoma cell line CNE-1. Cancer Biomark 22, 641–648 (2018).
Coomans de Brachene, A. & Demoulin, J. B. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 73, 1159–1172 (2016).
Zhao, M. et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat. Commun. 7, 11309 (2016).
Zheng, M. et al. STAT3 promotes invasion and aerobic glycolysis of human oral squamous cell carcinoma via inhibiting FoxO1. Front. Oncol. 9, 1175 (2019).
Jiang, G. et al. Role of STAT3 and FOXO1 in the divergent therapeutic responses of non-metastatic and metastatic bladder cancer cells to miR-145. Mol. Cancer Ther. 16, 924–935 (2017).
He, H. et al. MicroRNA-494-3p promotes cell growth, migration, and invasion of nasopharyngeal Carcinoma by Targeting Sox7. Technol. Cancer Res. Treat. 17, 1533033818809993 (2018).
Yin, W., Shi, L. & Mao, Y. MiR-194 regulates nasopharyngeal carcinoma progression by modulating MAP3K3 expression. FEBS Open Bio 9, 43–52 (2019).
Kawagoe, K. et al. Regulation of aberrantly expressed SERPINH1 by antitumor miR-148a-5p inhibits cancer cell aggressiveness in gastric cancer. J. Hum. Genet. https://doi.org/10.1038/s10038-020-0746-6 (2020).
Wang, H. et al. Radiofrequency ablation triggers the migration of hepatocellular carcinoma cells by suppressing miR-148a-5p. Biol. Chem. https://doi.org/10.1515/hsz-2020-0130 (2020).
Heo, M. J. et al. Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression. Gut 68, 708–720 (2019).
Su, B. et al. Profiling and comparing transcription factors activated in non-metastatic and metastatic nasopharyngeal carcinoma cells. J. Cell Biochem. 109, 173–183 (2010).
Zhang, Z. et al. SRGN-TGFbeta2 regulatory loop confers invasion and metastasis in triple-negative breast cancer. Oncogenesis 6, e360 (2017).
Guo, J. Y. et al. Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner. Oncogene 36, 2457–2471 (2017).
Bois, P. R. et al. FOXO1a acts as a selective tumor suppressor in alveolar rhabdomyosarcoma. J. Cell Biol. 170, 903–912 (2005).
Wei, Y. T., Guo, D. W., Hou, X. Z. & Jiang, D. Q. miRNA-223 suppresses FOXO1 and functions as a potential tumor marker in breast cancer. Cell Mol. Biol. 63, 113–118 (2017).
Aishanjiang, A. et al. MicroRNA-9 enhances invasion and migration of cervical carcinomas by directly targeting FOXO1. Eur. Rev. Med. Pharmacol. Sci. 22, 2253–2260 (2018).
Li, W. et al. miR-29c plays a suppressive role in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway. Clin. Epigenetics 10, 64 (2018).
Zheng, M. et al. EZH2 promotes invasion and tumor glycolysis by regulating STAT3 and FoxO1 signalling in human OSCC cells. J. Cell Mol. Med. 23, 6942–6954 (2019).
Acknowledgements
We are very grateful to Professor Sai-Wah Tsao from University of Hong Kong for generously presenting the NPC cell line C666-1 and normal immortalized nasopharyngeal epithelial cell line (NP69), Professor Musheng Zeng (Sun Yat-sen University Cancer Center) for kindly providing SUNE-1 and 5-8F cells. This research was supported by Guangxi Key Research and Development Plan (GuiKe-AB18050011), Guangxi Science and Technology Base and Talent Project (GuiKe-AD20297069), Youth Fund of the People’s Hospital of Guangxi Zhuang Autonomous Region (Code: QN2017-03), Open Research Project of Key Laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (GKE2d9-KF01).
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.L.W. and S.H.Q.; Methodology, D.R. and Y.L.W.; Investigation, J.Z.W. H.J. and J.L.L.; Writing – Original Draft, D.R.; Writing –Review & Editing, Y.L.W.; Funding Acquisition, J.L.; Resources, F.L.; Supervision, Y.L.W. and S.H.Q. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This research was approved by the Ethics Committee of the People’s Hospital of Guangxi Zhuang Autonomous Region, and informed consent was supplied by every subject involved in this study (No. 2018-47).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, YL., Ren, D., Lu, JL. et al. STAT3 regulates SRGN and promotes metastasis of nasopharyngeal carcinoma through the FoxO1-miR-148a-5p-CREB1 axis. Lab Invest 102, 919–934 (2022). https://doi.org/10.1038/s41374-022-00733-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-022-00733-7