Abstract
The mitochondrial intracrine Stanniocalcin 1 (STC1) activates mitochondrial anti-oxidant defenses. LRP2 (megalin) shuttles STC1 to the mitochondria through retrograde early endosome-to-Golgi- and Rab32-mediated pathway, and LRP2 KO impairs mitochondrial respiration and glycolysis. We determined STC1-LRP2 interaction domains using HA- and FLAG-tagged fragments of STC1 and LRP2, respectively, co-expressed in HEK293T cells. The trans-membrane domain of LRP2 is required for trafficking to the mitochondria. STC1-FLAG expressed in LRP2 KO cells fails to reach the mitochondria; thus, mitochondrial STC1 is extracellularly-derived via LRP2-mediated trafficking. Tri-leucines L12-14 in LRP2’s signal peptide interact with STC1’s signal peptide. Mutant LRP2 (L(12–14)A) does not bind STC1, while hSTC1 lacking signal peptide or Leucines L8/9/11 does not bind LRP2. STC1 fails to induce respiration or glycolysis in megalin KO mouse embryonal fibroblasts (MEF) expressing mutant LRP2, while mutant hSTC1 (L8/L9/L11 - > A8/A9/A11) fails to reach the mitochondria or induce respiration and glycolysis in WT MEF. Our data suggest direct regulation of mitochondrial metabolism by extracellular cues and reveal an important role for signal peptides’ leucines in protein–protein interactions and mitochondrial biology.
Similar content being viewed by others
Introduction
Mammalian stanniocalcins are secreted phospho-glycoproteins, that possess autocrine/paracrine actions1. STC1 is a mitochondrial intracrine [i.e., extracellular signaling molecule, targeted to the mitochondria2]; STC1 activates the metabolic sensor AMPK, which in turn upregulates uncoupling protein 2 (Ucp2) and Sirt33, to promote mitochondrial anti-oxidant defenses4,5. Transgenic overexpression of STC1 confers resistance to ischemia/reperfusion kidney injury4, while conditional and kidney-specific knockdown of STC1 increases ROS and leads to AKI5. Moreover, STC1 is a critical component of hypoxic preconditioning in the heart6 and brain7, and has been shown to mediate the protective actions conferred by mesenchymal stem cells8,9,10.
STC1 does not contain mitochondrial targeting sequence (based on ExPASy-PROSITE analysis) and thus, the mechanism of mitochondrial targeting is unknown. Indirect evidence suggests the existence of STC1 receptors at the plasma membrane and mitochondria11; however, the identity of the receptor/binding protein is also unknown. We have recently identified and characterized a megalin-dependent pathway for internalization and targeting of STC1 from the cell surface to the mitochondria via retrograde early endosome-to-Golgi- and Rab32-mediated trafficking12. In the mitochondria, STC1 exists in complex with megalin and SIRT312. This pathway mediates shuttling and trafficking of other mitochondrial intracrines such as angiotensin II and TGF-β12.
In the current work, we sought to characterize domains that mediate the interaction between STC1 and megalin, and define the contributions of megalin and STC1 to mitochondrial function. Our data suggest that megalin-mediated shuttling of extracellularly derived STC1 to the mitochondria regulates mitochondrial respiration and glycolysis, consistent with direct regulation of mitochondrial metabolism by extracellular cues. Signal peptides’ leucines-mediated interactions between STC1 and megalin play a critical role in mitochondrial biology. Of note, mutations in megalin have been linked to the pathogenesis of many diseases including Donnai-Barrow and Lowe syndromes13,14, and our data shed some light on possible shared pathway for to these diseases.
Methods
Cells and reagents
Human embryonic kidney HEK293T were cultured in DMEM medium with 10% FBS (Omega Scientific, Inc.). Primary WT and Lrp2 KO MEFs were generated from embryos of a Lrp2 + /− female mouse mated with Lrp2 + /− male mouse at 12.5 days postcoitum and cultured as described previously15. The DMEM medium (catalog #11965092) and MitoTracker™ Deep Red FM (M22426) were purchased from Thermofisher Scientific. The antibodies anti-ATP5B (sc-55597) and anti-vinculin (sc-25336) were obtained from Santa Cruz Biotechnology; anti-FLAG-HRP (A8592) and anti-HA-HRP (H6533) were purchased from Sigma.
Generation of recombinant expression constructs
STC1 and LRP2 deletion and mutation variants were generated by cloning or using the Q5® site-directed mutagenesis kit (New England Biolabs E0554S) based on wild type human LRP2 (Origene RC213707) or wild type human STC1 cDNA as previously described12. FLAG tag was added to the C-terminus of LRP2 variants. FLAG or hemagglutinin (HA)-tags were added to STC1 variant C-termini. Variant sequences for both LRP2 and STC1 were subsequently cloned into the pcDNA™3.1 mammalian expression vector (Invitrogen, V790-20). Wild type sequence validation, and deletion/mutation variant sequence comparisons and alignments were performed with DNAman software (Lynnon BioSoft). Sequencing analyses were performed by LoneStar Labs. Cloning oligonucleotides are listed in Supplementary Table 1.
In vitro transfection
For HEK293T cells, transient transfection was performed using Lipofectamine™ 3000 Transfection Reagent (ThermoScientific, L3000015) following the manufacturing instruction. MEF cells were electroporated with Neon transfection system at 1350 Voltage/30 width/1 shot, then seeded on dishes and cultured O/N before the next treatment.
Expression and purification of STC1-HA and STC1-FLAG fusion proteins
STC1-HA protein was purified from lysates of HEK293T cells transfected with wild type human STC1-HA using monoclonal anti-HA-agarose antibody (Sigma–Aldrich A2095). Briefly, anti-HA-agarose particles were added to the cell lysate and incubated overnight on an orbital shaker at 4 °C. Centrifuged resin was washed with phosphate-buffered saline four times, and STC1-HA protein was disassociated from agarose with 0.1 M glycine-HCl (pH 2.5) and then neutralized with 1 M Tris buffer (pH 8.0). STC1-FLAG, similarly overexpressed in HEK293T cells, was isolated using Anti-FLAG M2 magnetic beads (Sigma–Aldrich M8823) following the manufacturer instructions. Briefly, cell lysates were added to anti-FLAG magnetic beads and rotated overnight at 4 °C. Beads were isolated in magnetic separator (to collect the beads and remove the supernatant), washed 3 times with 20x volumes (of the packed magnetic beads) of TBS. The bound protein is eluted with 0.1 M glycine HCl, pH 3.0, followed by neutralization with 1 M Tris buffer, pH 8.0.
Protein preparation, immunoprecipitation and immunoblotting
For preparation of protein extracts from cultured cells, 25% of the cells are used for preparation of whole-cell protein extract and 75% of cells are used for preparation of mitochondrial protein and microsomal fractions using Mitochondria Isolation Kit for Cultured Cells (ThermoScientific, 89874) following the manufacturer’s instruction. For IP analysis, cell protein is prepared in low salt lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1.5 mM MgCl2, 1% Triton-X100); 300 µg protein are incubated with anti-FLAG M2 magnetic beads (M8823, Sigma) or anti-HA-agarose (A2095, Sigma) for 2 h at 4 °C, then washed three times, mixed with 3X SDS/PAGE loading buffer and boiled before loading onto SDS-gel. Microsomal (60 µg) or mitochondrial (40 µg) protein preparations are boiled in SDS sample buffer for 5 min, then loaded directly onto SDS gel, transferred onto nitrocellulose membranes (Bio-Rad, 10484059), blocked with 5% milk and probed with specific primary and then with goat anti-mouse or goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase. The blots are developed using the Supersignal West Dura Extended Duration Substrate (ThermoScientific, 34075) and images were captured using ChemiDoc MP Image System (BioRad, 17001402).
Confocal deconvolution fluorescence microscopy
Protein mitochondrial location was studied using confocal deconvolution fluorescence as described previously12. HEK293T cells seeded on micro-cover glasses were transiently transfected with corresponding plasmids and cultured overnight. MEF cells were electroporated with corresponding plasmids using Neo transfection system and cultured on glass disc overnight. For STC1-FLAG/STC1-HA internalization/targeting study, cells seeded on discs were treated with purified STC1 protein at 100 ng/mL (added to the medium) for 8 h. For mitochondrial localization study, cells were treated with 200 nM MitoTracker™ Deep Red FM for 30 min, then fixed in cold methanol at −20 °C for 10 min, then washed 3 times in PBS (5 min each), blocked in PBS containing 1% BSA for 1 h. The cells were then probed with specific primary antibody for 1 h at room temperature, washed 3 times in 1X PBS (5 min each), incubated with secondary antibodies conjugated with FITC or Alexa Fluor 405 (A-31556, ThermoScientific) for 1 h at room temperature in the dark. The cells were washed 3 times in PBS (5 min each) then mounted in DAPI-containing medium (H1200, VectorLaboratories) or without DAPI-containing medium. Images were captured using GE Healthcare DeltaVision live epifluorescence image restoration microscope installed with standard filter set for DAPI/FITC/TRITC/Cy5 and 60 × oil Olympus Plan Apo/Na1.4 objective lens and a 1.9 k × 1.9 × s CMOS camera.
Seahorse XF cell Mito-stress assay
Mitochondria respiration was examined using Seahorse XF Cell Mito Stress Test Kit (Agilent Technologies, North Billerica, MA) as previously described (12). Cells are seeded on XF cell culture microplate to 3 × 104 cells/per well density in 500 µl of culture medium. 50 mL of assay medium is prepared by supplementing Seahorse XF Base Medium with pyruvate (1 mM), glutamine (2 mM), and glucose (10 mM), adjusted to pH 7.4 with 0.1 N NaOH, followed by sterile filtration. A pre-hydrated sensor cartridge (pre-loaded with 1 mL of Seahorse XF calibration buffer and placed overnight at 37 °C in a non-CO2 incubator) is loaded with the following reagents which are freshly suspended in assay medium: Oligomycin (56 µL of 10 µM solution) to port A; carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP; 62 µL of 20 µM solution) to port B; Rotenone/Antimycin A (69 µL of a 5 µM solution) to port C, and mounted on the analyzer. Concomitantly, cell culture microplate is removed from 37 °C CO2 incubator, washed twice in assay medium, and incubated in 500 µL assay medium at 37 °C in non-CO2 incubator for 1 h prior to the assay. The calibration plate is replaced with the cell culture microplate at the start of the assay program. The original data are normalized to total protein, calibrated to non-mitochondrial respiration and statistically analyzed (n = 3).
Seahorse XF Glycolysis stress assay
Glycolysis is examined using Seahorse XF Glycolysis Stress Test Kit (Agilent Technologies, North Billerica, MA) as previously described (12). Cells are seeded in XF cell culture microplate to 3 × 104 cells/per well density in 500 µL of culture medium. 50 mL of assay medium are prepared by supplementing Seahorse XF Base Medium with glutamine (1 mM), adjusted to pH 7.4 with 0.1 N NaOH, followed by sterile filtration. A pre-hydrated sensor cartridge (pre-loaded with 1 mL of Seahorse XF calibration buffer, placed overnight at 37 °C in a non-CO2 incubator) is loaded with the following reagents which are freshly suspended in assay medium: Glucose (56 µL of 100 mM solution) to port A; Oligomycin (62 µL of 10 µM solution) to port B; 2-Deoxy-D-glucose (2-DG; 69 µL of a 500 mM solution) to port C, and mounted on the analyzer. Concomitantly, cell culture microplate is removed from 37 °C CO2 incubator, washed twice in assay medium, and incubated in 500 µL assay medium at 37 °C in non-CO2 incubator for 1 h prior to the assay. The calibration plate is replaced with the cell culture microplate at the start of the assay program. The original data are normalized to total protein, calibrated to the non-glycolytic acidification, and statistically analyzed (n = 3).
Statistical analysis
Data are represented as the mean ± standard deviation (STD). Differences between groups are analyzed using the Student’s t test. A P ≤ 0.05 is considered statistically significant.
Results
Transmembrane domain is required for megalin targeting to the mitochondria
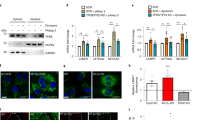
The open reading frame for human megalin mRNA encodes 4612 aa16. The primary structure of megalin predicts a type I transmembrane protein containing 25 aa N-terminal signal peptide, a 4398 aa extracellular region, a 23 aa single transmembrane domain, and a 209 aa C-terminal cytoplasmic tail16. To determine domain(s) responsible for megalin targeting to the mitochondria, we expressed in HEK293T cells, full-length hLRP2 (RC213707, Origene Technologies, Rockville, MD) and 5 overlapping megalin fragments spanning the entire aa sequence (all FLAG-tagged on the C-terminus) and determined their presence in the mitochondria. Fragment 1 spans N-terminal aa 1–849; fragment 2 spans N-terminus aa 775–1882; fragment 3 spans N-terminus aa 1819–2448; fragment 4 spans aa 2406–3778; fragment 5 spans C-terminal aa 3689–4612, which contains the transmembrane domain (Fig. 1A).
A Constructs of hLRP2 fragments. B–C Mitochondrial localization of products of different LRP2 constructs over-expressed in HEK293T cells reveals involvement of the C-terminus of LRP2 in mitochondrial localization by WB (B) and confocal deconvolution microscopy (C). Total cell (T), mitochondrial (M) and cytosolic (C) fractions. D Constructs of C-terminus LRP2 fragment hLRP2(3689–4612)-FLAG and hLRP2(3689–4612)ΔTM-FLAG (missing 29 aa containing putative 23 aa transmembrane domain designated TM: ALAIAGFFHYRRTGSLLPALPKLPSLSSL). E–H Deletion of 29 aa TM diminishes targeting of LRP2 C-terminal fragment to the mitochondria by WB (E–F) and confocal deconvolution microscopy (G–H). *p < 0.05.
Only the full-length hLRP2 and the C-terminal fragment (aa 3689–4612) were detected in the mitochondrial preparations (Fig. 1B). Their mitochondrial location was further verified by deconvolution microscopy (Fig. 1C). These data suggest that a domain contained within the C-terminus 924 aa is responsible for mitochondrial targeting of megalin; this fragment is predicted to contain 692 aa extracellular juxta-membrane region, a single 23 aa transmembrane domain and 209 aa cytosolic domain16. Endocytosis of megalin occurs in clathrin-coated pits17, and we reasoned that the trans-membrane domain serves as a membrane anchor for megalin. Indeed, deletion of 29 aa sequence (aa 4396–4424) that contains this domain diminishes megalin’s presence in the mitochondria, consistent with a requirement for the transmembrane domain for mitochondrial targeting of megalin (Fig. 1D–H).
STC1 binds to N-terminal leucines 12–14 of megalin
The extracellular region of megalin contains 3 types of cysteine-rich repeat motifs: 36 ligand-binding motifs (class A), 1 EGF motif (class B.1), and 15 growth factor motifs (class B.2)16. The 36 class A motifs are arranged in 4 clusters and, with the exception of the extreme N-terminus cluster, each class A cluster is flanked by a single B.2 motif at its N-terminus and two B.2 motifs at its C-terminus16. The remaining 4 B.2 motifs are contained within the YWTD repeat spacer regions that separate the four class A/B.2 clusters. A single B.1 motif is located in the extracellular juxtamembrane region. The cytoplasmic tail contains two coated-pit mediated internalization signal [F(X)NPXY], and several SH3 and SH2 motifs for signaling molecules16.
To determine domains within megalin that are responsible for STC1 binding, we co-expressed full-length hSTC1-HA with full-length hLRP2/megalin or full-length hSTC1-HA with one of the megalin fragments (Fig. 2A). The extreme N-terminus A cluster of megalin is contained within the first megalin fragment (aa 1–849), while the second megalin A cluster is contained within the second megalin fragment (aa 775–1882)16. Immunoprecipitation studies show STC1 associates with the full-length- and the N-terminus fragment of megalin (aa 1–849) (Fig. 2B, C), but not the other megalin fragments (Fig. 2D–G); thus, STC1 binds to a ligand binding motif within the first A cluster.
A Constructs of hLRP2 fragments. Full-length hLRP2 or fragments thereof are tagged with FLAG on the C-terminus. B–G Plasmids expressing full-length hLRP2 or LRP2 fragments were co-expressed with human STC1-HA (C-terminus HA-tag) plasmid in HEK293T cells. Immunoprecipitates with anti-FLAG antibody were performed. Whole cell lysates (WCL) or anti-FLAG immunoprecipitates were run on SDS-PAGE (1–12% gradient gel) and Western blots reacted with anti-FLAG and anti-HA. The interaction results are shown in panel (A).
Using the N-terminal fragment of megalin (aa 1–849) for additional sub-cloning, co-expression with STC1-HA, and immunoprecipitation/blotting, we narrowed down the binding site for STC1 to the N-terminal 100 aa of megalin (Fig. S1). Next, using the extreme N-terminus 221 aa fragment of megalin, we carried out progressive deletions of 25 aa, identifying the binding site for STC1 within the signal peptide of megalin (first 25 N-terminal aa of megalin; Fig. S2). Using the same 221 aa fragment of megalin, we then carried out progressive deletions of 5 aa at a time starting with aa 2 and found the deletion of aa 12–16 (LLLAL) within the signal peptide of megalin abolishes STC1 binding (Fig. S3); of note the aa motif LLLAL is conserved across evolution in mammalian megalins (Fig. 3A). Substitution of N-terminal 12–16 aa (LLLAL) or tri-leucine 12–14 with alanine is sufficient to abolish STC1 binding to the megalin fragment; however, substitution of a single leucine among L12-L14 does not affect the binding (Fig. 3B), while substitutions of 2 among the 3 leucines partially decreases STC1 binding to megalin (Fig. S4). Finally, we carried out substitution of tri-leucines L12-L14 to A12-A14 in the full-length megalin – to generate mutant megalin [LRP2 L(12–14)A] that does not bind STC1 (Fig. 4B). This mutant megalin is detected in the mitochondria upon overexpression in HEK293T cells, indicating that tri-leucine mutations does not affect megalin’s targeting to the mitochondria (Fig. 4C, D).
A N-terminal aa 12–16 (LLLAL) of megalin have been conserved across evolution in mammals. B cDNAs expressing N-terminal fragment of megalin (aa 1–221) or fragments thereof (all C-terminus FLAG-tagged) where the designated amino acid(s) was/were substituted with alanine, were expressed in HEK293T cells with or without hSTC1-HA (C-terminus HA-tag); whole cell (WCL) lysates or immunoprecipitates with either anti-HA or anti-FLAG were run on SDS-PAGE (12%) and Western blots reacted with anti-FLAG and anti-HA. The interaction results are shown in panel (B).
A Sequence validation of L12-L14 to A12-A14 mutation in full-length megalin. B Mutant megalin (LRP2m) fails to associate with STC1. Plasmids expressing FLAG-tagged WT megalin and mutant megalin (LRP2m, where signal peptide tri-leucines L12-L14 were substituted with alanines) were co-expressed with STC1-HA (C-terminus HA tag) in HEK293T cells; whole cell (WCL) lysates or immunoprecipitates with either anti-HA or anti-FLAG were run on SDS-PAGE (1–12% gradient gel) and Western blots were reacted with antibodies for FLAG and HA. C Mutation of megalin L(12–14)A does not affect its targeting to mitochondria. Mutant megalin was expressed in HEK293T cells; whole-cell and mitochondrial fractions were run on SDS-PAGE and Western blots were reacted with antibodies for FLAG, ATP5B (mitochondrial marker) and vinculin (microsomal marker). D Mitochondrial localization of mutant megalin is visualized using deconvolution confocal microscopy.
Mitochondrial STC1 is extracellularly-derived via megalin-mediated trafficking
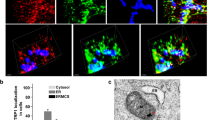
Mammalian stanniocalcins are secreted phosphoglycoproteins1, and published work from our lab shows extracellularly applied STC1 is targeted to the mitochondria in WT C2C12 cells12; but fails to reach the mitochondria in Lrp2 KO C2C12 cells12. ExPASy-PROSITE analysis of STC1 protein predicts no mitochondrial targeting sequence; therefore, we examined the hypothesis that mitochondrial STC1 is extracellularly derived via megalin-mediated trafficking. We expressed full-length hSTC1-FLAG in WT and Lrp2 KO MEF cells. As shown in Fig. 5A, when expressed in WT MEF cells, hSTC1-FLAG is detected in the medium, microsomal and mitochondrial fractions. On the other hand, when expressed in Lrp2 KO cells, hSTC1-FLAG is detected in the microsomal fraction and medium, but not mitochondria (Fig. 5A). Thus, STC1-FLAG is expressed and secreted to the medium in both WT and Lrp2 KO MEF cells, but fails to reach the mitochondria in Lrp2 KO MEF cells. Next, we expressed WT and mutant human megalin [LRP2 L(12–14)A] in Lrp2 KO MEF cells, and applied hSTC1-HA protein extracellularly; STC1-HA is detected in whole-cell and mitochondrial fractions of Lrp2 KO MEF-transfected with WT megalin, but not in Lrp2 KO MEF-transfected with LRP2 L(12–14)A (Fig. 5B, C). Collectively, our data suggest that mitochondrial STC1 is extracellularly-derived via megalin-mediated trafficking, and substitution of leucines 12–14 in megalin abolishes trafficking of hSTC1 from the cell surface to the mitochondria.
A Full-length STC1-FLAG fails to reach the mitochondria when expressed in megalin KO mouse embryonic fibroblasts (MEF). Plasmid expressing full-length FLAG-tagged hSTC1 was transfected into WT or Lrp2 KO MEFs. Mitochondrial preparations (MP), microsomal fractions (MIC) and medium were run on SDS-PAGE, followed by blotting with antibodies for FLAG, ATP5B (mitochondrial marker) and vinculin (microsomal marker). B Extracellularly applied STC1-HA protein fails to reach the mitochondria when applied to Lrp2 KO MEF expressing mutant megalin (LRP2m). Lrp2 KO MEF cells were transfected with LRP2m-FLAG-expressing plasmid, followed by application of STC1-HA protein to the medium. Whole-cell and mitochondrial fractions were run on SDS-PAGE (1–12% gradient gel), followed by immunoblotting with antibodies for HA, FLAG, ATP5B (mitochondrial marker) and vinculin (microsomal marker). C Deconvolution confocal microscopy shows STC1 fails to enter the cell or reach the mitochondria when applied to Lrp2 KO MEF expressing mutant megalin.
Megalin interacts with STC1 via its signal peptide
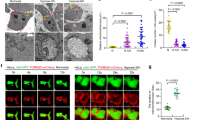
To determine STC1 domains that interact with megalin, we co-expressed in HEK293T cells, full-length hLRP2/megalin (C-terminus FLAG-tagged) with full-length hSTC1 [hSTC1 is 247 aa protein18] or fragments thereof (all C-terminus HA-tagged), as depicted in Fig. 6A. Immunoprecipitation studies reveal association between megalin and full-length STC1, as well as N150 and N200, but not fragments lacking 50- or 100-N-terminal aa of STC1 (Fig. 6A, B). When applied to the extracellular milieu of HEK293T cells, full-length hSTC1 is detected in the mitochondria, while hSTC1 lacking N-terminal 49 aa [hSTC1Δ(2–50)N-FLAG] does not (Fig. 6C–E). Secondary structure analysis for hSTC1 predicts N-terminal signal peptide (aa 1–17) and a pro-peptide (aa 18–33), while the mature peptide contains 214 aa19. To further characterize STC1 domain(s) that interact(s) with megalin, we generated hSTC1 constructs with progressive N-terminal deletions starting with aa 2, ending with aa 44, as shown in Fig. 6F. Co-expression and immunoprecipitation studies indicate interaction between megalin and the signal peptide of hSTC1 (Fig. 6G). Leucine repeats are over-represented in the signal peptides of secreted and type I membrane proteins of mammals, suggesting that leucines may have important and yet to be defined functions20; therefore, we hypothesized that leucines within the signal peptide of STC1 may be important for the interaction with megalin. We generated hSTC1 expressing constructs with deletions of aa 2–7 or aa 8–17 (contains 3 leucines), and a construct expressing mutated L8/L9/L11 to A8/A9/A11 (Fig. S5A). These mutants were confirmed with sequencing analysis (Fig. S5B). Immunoprecipitation studies reveal interaction between the megalin fragment and full-length hSTC1 only; the remaining constructs expressed truncated proteins (faster migrating relative to full-length hSTC1) that did not bind the megalin fragment (Fig. S5C); these peptides do not reach the mitochondria when applied to extracellular milieu of HEK293T cells (Fig. S5D). Furthermore, L8/L9/L11 to A8/A9/A11 mutations led to the expression of a truncated protein similar in size (migration) to hSTC1 lacking the signal peptide (Fig. S6A); both proteins are not secreted to the medium (Fig. S6B), suggesting that L8/L9/L11 to A8/A9/A11 mutation led to cleavage of the signal peptide, which is required for secretion of STC1. Importantly, secreted hSTC1 is equivalent in size (migration) to the full-length hSTC1 isolated from the intracellular fraction, suggesting that the signal peptide is not cleaved upon secretion of hSTC1 to the medium (Fig. S6C).
A Depiction of full-length hSTC1 and fragments thereof (all C-terminus HA-tagged). B Plasmids expressing FLAG-tagged N-terminus fragment of megalin (aa 1–849) and HA-tagged full-length hSTC1 or fragments thereof, were transfected into HEK293T cells. Whole-cell (WCL) lysates were subjected to immunoprecipitation using anti-HA. WCL and immuno-precipitates were run on SDS-PAGE (10%), and Western blots were reacted with anti-FLAG and anti-HA. The interaction results are shown in panel (B). C–D HEK293T cells were treated with vehicle (PBS), full-length STC1-FLAG or hSTC1Δ(2–50)N-FLAG. Extracellularly applied hSTC1-FLAG is detected in the mitochondria; but not hSTC1Δ(2–50)N-FLAG. STC1 localization was studied by both Western blot loaded with mitochondrial preparations (MP) reacted with anti-FLAG, or anti-Atp5B (mitochondrial marker) and deconvolution confocal. E Statistical analysis of mitochondrial localization of extracellularly applied STC11-FLAG or hSTC1Δ(2–50)N-FLAG. F Depicts N-terminal aa deletions in hSTC1. G Plasmids expressing FLAG-tagged N-terminus fragment of megalin (aa 1–143) and full-length hSTC1 or hSTC1 with deletions as depicted in panel (F) (all C-terminus HA-tagged), were transfected into HEK293T cells. Whole-cell lysates (WCL) were subjected to immunoprecipitation using anti-HA or anti-FLAG. WCL and immunoprecipitates were run on SDS-PAGE (10%), and Western blots were reacted with anti-FLAG and anti-HA. The interaction results are shown in panel (G). Data show LRP2 interacts with the signal peptide of hSTC1.
Megalin-mediated targeting of STC1 to the mitochondria is required for regulation of respiration and glycolysis
Published work from our lab shows megalin mediates shuttling of extracellularly applied full-length hSTC1 to the mitochondria, and megalin KO diminishes mitochondrial respiration and glycolysis12. We sought to determine whether mitochondrial targeting of STC1 is required for the regulation of mitochondrial function. As shown in Fig. S5, mutant-hSTC1 (L8/L9/L11 to A8/A9/A11) does not bind megalin. In wild type WT MEF, full-length hSTC1 protein stimulates basal and maximal respiration, ATP generation and spare respiratory capacity, while mutant hSTC1 does not (Fig. 7A). As we previously reported in Lrp2 KO C2C12 cells12, Lrp2 KO MEF cells show diminished basal and maximal respiration, ATP generation and spare respiratory capacity; neither full-length hSTC1 nor mutant-hSTC1 altered these responses (Fig. 7A). Similarly, in WT MEF, full-length hSTC1 stimulates baseline glycolysis, glycolytic capacity and glycolytic reserve, but mutant-hSTC1 does not (Fig. 7B). As we previously reported in Lrp2 KO C2C12 cells12, Lrp2 KO MEF show diminished baseline glycolysis, glycolytic capacity and reserve, and these responses were not altered by full-length hSTC1 or mutant-hSTC1 (Fig. 7B).
A Wild type (WT) or megalin/Lrp2 KO mouse embryonal fibroblasts (MEF) were treated with either full-length hSTC1 or hSTC1m. Baseline and maximal mitochondrial respiration, ATP production and spare respiratory capacity are induced by full-length hSTC1, but not hSTC1m (n = 3). Basal and maximal respiration, as well as spare capacity and ATP production were reduced in megalin KO MEF cells; full-length STC1 and hSTC1m fail to change these values (n = 3). B Similarly, in WT MEFs, baseline glycolysis, glycolytic capacity and glycolytic reserve were stimulated by full-length hSTC1, but not hSTC1m (n = 3). In megalin KO MEFs, baseline glycolysis, glycolytic capacity and glycolytic reserve were diminished; full-length STC1 and mutant STC1 fail to alter these results. Non-glycolytic acidification was not altered by full-length STC1 or hSTC1m in either WT or megalin KO MEFs (n = 3).
In the second set of experiments, we sought to determine the effects of full-length hSTC1 on mitochondrial respiration and glycolysis in Lrp2 KO MEFs-transfected with either WT or mutant megalin [LRP2-L(12–14)A, that does not bind hSTC1 (Fig. 4B)]. In WT megalin-transfected MEF, full-length hSTC1 stimulated baseline and maximal respiration, spare respiratory capacity and ATP generation; however, it failed to do so in mutant megalin-transfected cells (Fig. 8A). Similarly, in WT megalin-transfected Lrp2 KO MEF, hSTC1 increased baseline glycolysis, glycolytic capacity and glycolytic reserve; but failed to do so in mutant megalin-transfected Lrp2 KO MEF cells (Fig. 8B). Of note, Lrp2 KO MEFs and Lrp2 KO MEFs-transfected with mutant megalin displayed diminished and equivalent glycolytic profiles. Collectively, our data suggest that megalin-mediated internalization of STC1 to the mitochondria is important for the regulation of mitochondrial respiration and glycolysis.
A Megalin/Lrp2 KO MEFs were transfected with cDNAs that express either full-length WT hLRP2 or mutant human megalin [LRP2m; LRP2-L(12–14)A]. Cells were assayed 24 h later. Compared with Lrp2 KO MEFs, Lrp2 KO MEF-transfected with WT hLRP2 display higher baseline and maximal mitochondrial respiration, spare respiratory capacity and ATP production (n = 3). Baseline and maximal mitochondrial respiration, spare respiratory capacity and ATP production are stimulated by hSTC1 in megalin KO MEFs-transfected with WT megalin, but not in Lrp2 KO MEFs-transfected with LRP2m (n = 3). STC1 has no effect on respiration in Lrp2 KO MEFs (n = 3). B Lrp2 KO MEFs were transfected with cDNAs that express WT hLRP2 or mutant human megalin (LRP2m, which does not bind STC1). Cells were assayed 24 h later. Compared with Lrp2 KO MEFs, Lrp2 KO MEFs-transfected with WT hLRP2 displayed higher baseline glycolysis, glycolytic capacity and glycolytic reserve (n = 3). Baseline glycolysis, glycolytic capacity and glycolytic reserve were stimulated by STC1 in Lrp2 KO MEFs-transfected with WT hLRP2, but not in Lrp2 KO MEFs-transfected with hLRP2m (n = 3).
Discussion
The concept of mitochondrial intracrines refers to intracellularly-acting, extracellular signaling proteins/peptides2. Mitochondria represent one site of intracrine action, and among the intracrines present in the mitochondria include, TGF-β, GH, ANP, Wnt-13, STC1, angiotensin II and other renin-angiotensin system components2. We have shown megalin mediates trafficking to the mitochondria of STC1, angiotensin II and TGF-β12. Mitochondrial actions ascribed to angiotensin II include stimulation of mitochondrial nitric oxide production and modulation of respiration21; however, little is known about the effects on mitochondria of other mitochondrial intracrines. In this paper, we provide evidence that trafficking of the intracrine STC1 from the extracellular milieu to the mitochondria is important for the regulation of mitochondrial respiration and glycolysis, and the mitochondrial phenotype observed upon megalin deletion is attributable to abrogation of STC1 trafficking to the mitochondria12. This conclusion is based on the fact that mutant megalin with substitution of L12-L14 to A12-A14 does not bind STC1 (Fig. 4B) and is present in the mitochondria (Fig. 4C); while extracellularly applied STC1 is not detected in the mitochondria of mutant-megalin expressing cells (Fig. 5B). Thus, substitution of L12-L14 to A12-A14 does not impede megalin’s trafficking to the mitochondria and is not expected to interfere with megalin functions that are independent of STC1 binding/actions. With that in mind, L12-L14 to A12-A14 substitutions in megalin eliminate functions related to STC1/megalin interaction and trafficking of STC1 to the mitochondria. Indeed, current data suggest that trafficking of STC1 from the extracellular milieu to the mitochondria is required for the regulation of mitochondrial respiration and glycolysis, and interference with STC1 trafficking is responsible for the mitochondrial phenotype observed upon megalin deletion12. Thus, our data reveal an important role for megalin in mitochondrial biology, as it mediates a novel signaling pathway to regulate mitochondrial function – by direct trafficking of intracrine STC1 from the extracellular milieu to the mitochondria12. This novel pathway may be common to other mitochondrial intracrines, beyond STC1, angiotensin II and TGF-β2,12. Moreover, since STC1 lacks mitochondrial targeting sequence (based on ExPASy-PROSITE analysis), megalin-mediated trafficking of STC1 to the mitochondria may represent an example of a novel pathway to deliver other proteins lacking conventional mitochondrial targeting sequences to the mitochondria (e.g., angiotensin II, TGF-β, etc).
Also notable is the fact that the signal peptides of STC1 and megalin mediate their interaction. Signal peptides are found on proteins targeted for the secretory pathway and include proteins within organelles (e.g., ER, Golgi, endosomes), proteins inserted into cellular membranes and secreted proteins22,23,24. The common structure of signal peptides contains a positively charged n-region (1–2 aa long containing Lys or Arg), followed by a hydrophobic h-domain (average length of 12 aa and contains predominantly Val, Leu, Ile, Ala, Phe, Met and Trp) and a neutral but polar c-region (5–7 aa long) containing the peptidase cleavage site, which has a universally conserved “(−3, −1) design”25,26, where the residues at positions −3 and −1, relative to the cleavage site, must be small and neutral for correct cleavage to occur. Signal peptides are usually cleaved by signal peptidase in the ER; however, many are retained and fulfill functions beyond targeting of the peptide to its destination. Such is the case for STC1 and megalin, where the signal peptides in the secreted form of STC1 and the plasma membrane-anchored megalin, are preserved and serve as interacting domains. Of interest, tri-leucines L12-L14 within the signal peptide of megalin are responsible for STC1 binding; conversely, deletion of leucines L8/L9/L11 within the signal peptide of STC1 results in loss of the signal peptide, abolishing STC1 secretion and binding to megalin. A detailed analysis of aa repeats within signal peptides of type I membrane proteins and secreted peptides (e.g., megalin) of over 100 species reveals over-representation of leucine repeats (but not other hydrophobic aa) in signal peptides of higher eukaryotes, particularly mammals20. And while <1/5th of all proteins in the human proteome have signal peptides, nearly 2/3rd of all leucine repeats are found in signal peptides. Moreover, leucine repeats appear to have a variety of roles in sorting signals and intracellular trafficking. NPXY, YXXØ and dileucines serve as sorting signals in proteins, and proteins containing these sorting signals have different steady state distributions in the cell, localizing at the plasma membrane, recycling endosomes, late endosomes, lysosomes and the TGN, melanosomes and synaptic vesicles27,28. These sorting signals act as internalization signals when the proteins are at the plasma membrane, and help determine the trafficking itineraries and steady state distributions inside the cell29. Thus, conservation of the leucine-based signal peptides in both megalin and STC1 is noteworthy, and the critical role that leucines play in their interactions suggest that leucine—based signal peptides may have an important role in cell biology and mitochondrial signaling.
Neurological developmental abnormalities are observed in humans with autosomal recessive Donnai-Barrow and facio-oculo-acoustico-renal (DB/FOAR) syndrome where various loss-of-function mutations in LRP2 result in corpus callosum agenesis, as well as ocular, hearing, and facial abnormalities accompanied by low-molecular weight proteinuria13. In the closely related Lowe syndrome (oculocerebrorenal), characterized by congenital cataracts, cognitive impairment, proximal tubular dysfunction and arthropathy, megalin function depends upon an intact endosome-lysosome pathway. Lowe syndrome is an inherited X-linked disease that is caused by mutations in the inositol polyphosphate 5-phosphatase gene (OCRL) and associated with impaired Rab GTPase binding and clathrin regulation leading to dysfunctional megalin-mediated endocytosis14. Our data also suggest that the pathogenesis of Donnai-Barrow and Lowe syndromes may be linked to abnormal mitochondrial intracrine signaling.
Data availability
All data generated or analyzed during this study are included in this published article [and its Supplementary Information files].
References
Jellinek, D. A. et al. Stanniocalcin 1 and 2 are secreted as phosphoproteins from human fibrosarcoma cells. Biochem. J. 350, 453–461 (2000).
Re, R. N. & Cook, J. L. The mitochondrial component of intracrine action. Am. J. Physiol. Heart Circ. Physiol. 299, H577–H583 (2010).
Pan, J. S. et al. Stanniocalcin-1 inhibits renal ischemia/reperfusion injury via an AMP-activated protein kinase-dependent pathway. J. Am. Soc. Nephrol. 26, 364–378 (2015).
Huang, L. et al. Overexpression of stanniocalcin-1 inhibits reactive oxygen species and renal ischemia/reperfusion injury in mice. Kidney Int. 82, 867–877 (2012).
Huang, L. et al. AKI after conditional and kidney-specific knockdown of Stanniocalcin-1. J. Am. Soc. Nephrol. 25, 2303–2315 (2014).
Westberg, J. A., Serlachius, M., Lankila, P. & Andersson, L. C. Hypoxic preconditioning induces elevated expression of stanniocalcin-1 in the heart. Am. J. Physiol. Heart Circ. Physiol. 293, H1766–H1771 (2007).
Westberg, J. A. et al. Hypoxic preconditioning induces neuroprotective stanniocalcin-1 in brain via IL-6 signaling. Stroke 38, 1025–1030 (2007).
Oh, J. Y. et al. Mesenchymal stem/stromal cells inhibit the NLRP3 inflammasome by decreasing mitochondrial reactive oxygen species. Stem Cells 32, 1553–1563 (2014).
Ono, M. et al. Mesenchymal stem cells correct inappropriate epithelial-mesenchyme relation in pulmonary fibrosis using stanniocalcin-1. Mol. Ther. 23, 549–560 (2015).
Togel, F. et al. Administered mesenchymal stem cells protect against ischemic acute renal failure through differentiation-independent mechanisms. Am. J. Physiol. Renal. Physiol. 289, F31–F42 (2005).
McCudden, C. R., James, K. A., Hasilo, C. & Wagner, G. F. Characterization of mammalian stanniocalcin receptors. Mitochondrial targeting of ligand and receptor for regulation of cellular metabolism. J. Biol. Chem. 277, 45249–45258 (2002).
Li, Q. et al. Megalin mediates plasma membrane to mitochondria cross-talk and regulates mitochondrial metabolism. Cell Mol. Life Sci. 75, 4021–4040 (2018).
Kantarci, S. et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 39, 957–959 (2007).
De Matteis, M. A., Staiano, L., Emma, F. & Devuyst, O. The 5-phosphatase OCRL in Lowe syndrome and Dent disease 2. Nat. Rev. Nephrol. 13, 455–470 (2017).
Li, Q., Chu, M. J. & Xu, J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol. Cell Biol. 27, 8073–8086 (2007).
Hjalm, G. et al. Cloning and sequencing of human gp330, a Ca(2+)-binding receptor with potential intracellular signaling properties. Eur. J. Biochem. 239, 132–137 (1996).
Nielsen, R., Christensen, E. I. & Birn, H. Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int. 89, 58–67 (2016).
Olsen, H. S., Cepeda, M. A., Zhang, Q. Q., Rosen, C. A. & Vozzolo, B. L. Human stanniocalcin: a possible hormonal regulator of mineral metabolism. Proc. Natl Acad. Sci. USA 93, 1792–1796 (1996).
Trindade, D. M., Silva, J. C., Navarro, M. S., Torriani, I. C. & Kobarg, J. Low-resolution structural studies of human Stanniocalcin-1. BMC Struct. Biol. 9, 57 (2009).
Labaj, P. P., Leparc, G. G., Bardet, A. F., Kreil, G. & Kreil, D. P. Single amino acid repeats in signal peptides. FEBS J. 277, 3147–3157 (2010).
Abadir, P. M. et al. Identification and characterization of a functional mitochondrial angiotensin system. Proc. Natl Acad. Sci. USA 108, 14849–14854 (2011).
Blobel, G. & Dobberstein, B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67, 852–862 (1975).
Blobel, G. & Dobberstein, B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67, 835–851 (1975).
Martoglio, B. & Dobberstein, B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8, 410–415 (1998).
von Heijne, G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14, 4683–4690 (1986).
von Heijne, G. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133, 17–21 (1983).
Robinson, M. S. & Bonifacino, J. S. Adaptor-related proteins. Curr. Opin. Cell Biol. 13, 444–453 (2001).
Mattera, R., Boehm, M., Chaudhuri, R., Prabhu, Y. & Bonifacino, J. S. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J. Biol. Chem. 286, 2022–2030 (2011).
Bonifacino, J. S. & Dell’Angelica, E. C. Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 145, 923–926 (1999).
Funding
This work was supported by grants from: The Veteran Administration [DSH: 2I01BX002006; JS-CP: 1IK2BX002912], the American Society of Nephrology (MH), and a generous gift from Dr. and Mrs. Harold Selzman. This project was also supported by the Pathology and Histology Core at Baylor College of Medicine, with funding from the NIH (NCI P30-CA125123), and the expert assistance of Michael Ittmann, MD PhD. Imaging for this project was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from NIH (DK56338, and CA125123), CPRIT (RP150578), the Dan L. Duncan Comprehensive Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics.
Author information
Authors and Affiliations
Contributions
D.S.H.: conceptualization, formal analysis, funding acquisition, project administration, supervision, writing—original draft, writing—review & editing. Q.L.: conceptualization, data curation, formal analysis, investigation, methodology, writing—review & editing. S.-C.P.: formal analysis, writing—review & editing. L.T.: data curation, investigation. M.H.: data curation, formal analysis, investigation, methodology, visualization. J.L.: data curation, investigation. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Given by all authors.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Q., Holliday, M., Pan, J.SC. et al. Interactions between leucines within the signal peptides of megalin and stanniocalcin-1 are crucial for regulation of mitochondrial metabolism. Lab Invest 102, 534–544 (2022). https://doi.org/10.1038/s41374-022-00729-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-022-00729-3