Abstract
Breast fibroepithelial lesions (FEL) are biphasic tumors which consist of benign fibroadenomas (FAs) and the rarer phyllodes tumors (PTs). FAs and PTs have overlapping features, but have different clinical management, which makes correct core biopsy diagnosis important. This study used whole-slide images (WSIs) of 187 FA and 100 PT core biopsies, to investigate the potential role of artificial intelligence (AI) in FEL diagnosis. A total of 9228 FA patches and 6443 PT patches was generated from WSIs of the training subset, with each patch being 224 × 224 pixel in size. Our model employed a two-stage architecture comprising a convolutional neural network (CNN) component for feature extraction from the patches, and a recurrent neural network (RNN) component for whole-slide classification using activation values from the global average pooling layer in the CNN model. It achieved an overall slide-level accuracy of 87.5%, with accuracies of 80% and 95% for FA and PT slides respectively. This affirms the potential role of AI in diagnostic discrimination between FA and PT on core biopsies which may be further refined for use in routine practice.
Similar content being viewed by others
Introduction
Breast fibroepithelial lesions (FELs) comprise fibroadenomas (FAs) and the less commonly occurring phyllodes tumors (PTs). PTs are classified as benign, borderline and malignant according to criteria established by the World Health Organization1,2. It is crucial that the correct diagnosis is determined, especially on core biopsies, since FAs and PTs have different clinical management. Women with FAs are subjected to imaging surveillance or a simple enucleation, while those with PTs conventionally require surgical removal with clear margins. PTs are likely to be larger and have more rapid growth than FAs. Benign PTs are reported to have a local recurrence rate of 10–17%, while rates for borderline and malignant PTs are estimated at 14–25% and 23–30%, respectively1,3. Grade progression can also occur during PT recurrences. Diagnosing FELs can be a challenge, as PTs mimic FAs clinicoradiologically, and histological evaluation is necessary to distinguish the two lesions4,5. Cellular FA and benign PT share overlapping morphological features, leading to difficulties in discriminating between them, especially in core needle biopsies (CNB)6. CNB can be performed in an outpatient setting, is minimally invasive, and is a good diagnostic tool for pre-operative assessment of breast lesions7,8,9,10,11,12,13,14,15,16. However false-negative results can occur due to under-sampling and reflecting only a small part of the tumor, and may not adequately represent a heterogeneous tumor17,18. Well-reported issues of reproducibility and inter-observer variability add to the difficulties of achieving an accurate diagnosis12,19,20. Attempts for a more robust preoperative tool to aid morphological assessment in distinguishing between FAs and PTs have resulted in the creation of two assays, both tested on CNBs. A 5-gene reverse transcription-PCR assay (which measures the expression of ABCA8, APOD, CCL19, FN1 and PRAME)21 reported prediction accuracy rates of 94.7% (179/189) and 82.9% (34/41) for FAs and PTs respectively. Additionally, our group developed a 16-gene targeted next generation sequencing panel (FEB assay) to profile FELs22,23, having an accuracy of 89.6%, a specificity of 95.8%, and a sensitivity of 65.1%. Both assays still face the issue of discordant results, in which original pathology conclusions differed from predicted diagnoses. This was observed in 17 out of 230 cases for the 5-gene PCR test, while the FEB assay had 27 discordant cases out of 211 lesions profiled. For the latter, this was partly because of some FAs harboring a TERT promoter mutation and/or mutations in other genes such as RB1 and SETD222. Other factors which may be of concern in considering a genetic test include longer turnaround time, higher cost and sample quality, since formalin-fixed paraffin-embedded (FFPE) tissues have a greater likelihood of DNA degradation, low DNA yield, formalin crosslinking and sequencing artefacts. Given these limitations, there is a need to explore other platforms for improved diagnostics. In the current era of increasing integration of computational and digital pathology into routine diagnostics, a more specific diagnostic tool utilizing artificial intelligence (AI) that can detect FAs versus PTs effectively and quickly in CNBs is thus desirable, and we aim to investigate its potential application in this study.
Recent advances in whole-slide imaging have opened up the possibility to use computational methods to analyze histopathological features of biopsies captured in whole-slide images (WSIs), with particular interest in using AI, mostly involving deep learning methods, to assist in pathological classification and diagnosis. Convolutional neural networks (CNN) have shown promising results to extract relevant features for image analytics24. However, due to the large size of WSIs (few hundred megabytes to few gigabytes for typical 20×–40× images and pixel resolution in billions), direct application of CNN on WSIs is computationally challenging. Hence it is common for smaller image frames (sometimes known as “patches” or “tiles”) to be generated from WSIs as a way to reduce the dimension of the problem first before applying computational algorithms to classify each of the individual patches. This is followed by a second step of pooling/aggregating the patch-level outputs to derive slide/WSI level conclusions24,25,26. For the purposes of aggregating patch-level analysis into slide-level classification on histopathological WSIs, Iizuka et al.24 compared max pooling and recurrent neural networks (RNN), and found RNN to have reduced log-loss and less tendency for error. We used a similar two-stage model concept: applying CNN on smaller image patches to extract detailed features and classify each patch; followed by RNN to aggregate the patch-level outputs, taking into account their spatial relationships to produce the WSI/slide-level classification. Although there were past studies looking at analyzing image sets (e.g., smaller PNG images) or extracted smaller image frames from WSIs of breast tumors27,28,29,30, to date, we have not found similar studies analyzing breast FELs on WSIs exclusively from CNBs using deep learning methods.
Materials and methods
Data acquisition
Histology reports were reviewed during case selection, with diagnosis, patient age and specimen date recorded. Biopsies with any of the following descriptions in their reports were pulled out and used for the study: FA, cellular FA, hyalinized FA, PT, and FEL/cellular FEL which may favor FA or PT (Fig. 1). These were composed of core needle, trucut and mammotome biopsies.
A Light microscopy images at low magnification showing the core biopsy of a fibroadenoma with intracanalicular and pericanalicular patterns, and (B) its subsequent resection. C Benign phyllodes tumor biopsy, with (D) visible fronds in its resection specimen. E Biopsy of a borderline phyllodes tumor and (F) its resection displaying fronds and a cellular spindle cell stroma; moderate stromal nuclear pleomorphism and increased mitoses were discovered on the excision warranting a borderline grade.
Diagnoses of their subsequent resections (if available) were checked to confirm the final diagnoses and PT grade. Malignant PTs were not selected, as their histology can usually be readily distinguished from FAs on core biopsy. In addition, cases which were reported as FA in CNBs but PT in subsequent resections, that may occur due to under-sampling on CNB, were excluded. Those diagnosed as PT in CNBs but FA in excision were also excluded. A total of 187 FA and 100 PT (81 benign and 19 borderline) core biopsies was finally included in the study (Tables 1 and 2). Archival slides from 2011 to 2017 were obtained from the Department of Anatomical Pathology, Singapore General Hospital. Slides were checked by a pathologist. The slides of the selected core biopsies were scanned into WSIs using the Philips Ultra Fast Scanner (UFS) into Philips Intellisite Pathology Solution (PIPS). WSIs were then downloaded in iSyntax format from PIPS for further processing during algorithm training and analysis.
Stratified data split
The dataset was divided into three subsets, namely the training, validation and testing subsets, for the purposes of model development and evaluation. The training subset contained slides from which the model learnt parameters for distinguishing FAs and PTs. As part of the training process, these parameters were iteratively adjusted based on prediction performance evaluated on the validation subset. The testing subset was withheld from the training process, and used for the purpose of independently evaluating the final performance of the model on ‘unseen’ cases. Our testing subset comprised 40 slides selected from our full dataset by pathologists in the team. The remaining slides were then split into training and validation subsets of 197 and 50 slides, respectively. As patient age was hypothesized to be an important covariate in the characteristics of FELs1, the data split was performed in a randomized manner, and stratified based on patient age (Supplementary Fig. 1) to ensure that each of the data subsets was representative of the full dataset in terms of this factor. This is an important step in the machine learning workflow that helps to reduce potential bias arising from key attributes of the data (i.e., age) being different in the training subset versus the validation and testing subsets. Stratification was performed by categorizing patient age into 10-year age bands, and the data split was performed such that relative frequencies between age bands were preserved in the training and validation subsets. While patient age was used for data stratification, it was not included as an input feature to the model. In other words, the model was trained only on morphological features learnt from slide images.
Lesional annotations
Lesional regions within training and validation slides were annotated by the pathology team to aid in training the model to identify image patches containing features that are the most discriminative of FA or PT. Scanned images were downloaded from the PIPS as iSyntax files, converted into TIFF and uploaded onto the Open Microscopy Environment (OMERO) web-based platform. During the data labeling stage in OMERO, pathologists were asked to zoom in to the appropriate resolution level and identify the tissue region that is either FA or PT. A polygon was drawn by enclosing (with a margin as tight as possible) the entire region of lesional tissue. Specific cellular/subcellular features (e.g., epithelium vs stroma, nucleus vs cytoplasm) were not annotated. In the slides from the testing subset, lesional regions were not annotated, so as to better represent performance in deployment clinical use-cases where such annotations would not be available.
Patch generation and filtering
The extremely high resolution of the whole-slide images (~100,000 pixels width by 70,000 pixels height) imposed significant computational demands. It was therefore necessary to distribute the computational load by generating smaller, non-overlapping image patches from each whole-slide image. We used square patches of 224 pixels by 224 pixels in order to take advantage of convolutional neural network architectures pre-trained on images of the same dimensions (see model architecture section). Patches generated from whole-slide images were subject to a series of quality checks aimed at excluding patches unlikely to contain information that would aid in distinguishing FA and PT classes. Specifically, we aimed to exclude patches containing handling stains used in specimen preparation, imaging artifacts, and non-lesional tissue structures.
Ink stain detection
There were slides within the dataset that contained blue/green stains from handling dyes used during the biopsy process. For each slide, we generated a mask from pixels within the RGB value range of ink stains observed in the dataset. Based on this mask, patches containing ink stains were excluded.
Blur, background and slide edge detection
Image sharpness was determined from the variance of Laplacian operation outputs. Low variance indicated potentially out-of-focus images, which were excluded from the dataset. Slide background detection was implemented by calculating the proportion of pixels that exceeded light and dark thresholds. Images containing glass slide edges were detected using Hough line transformations and subsequently removed.
Folded tissue detection
Some slides contained folded (i.e., overlapping) tissue artifacts that could potentially impact model training. We carried out the folded tissue detection by first converting the image to the HSV color model and then taking the difference of saturation and luminance channel31. Large difference values are indicative of the increased dye absorption and thickness in regions containing folded tissue (i.e., high saturation, low luminance).
Adipose and blood detection
Patches containing adipocyte fragments were identified and excluded by finding image contours that were within range of typically observed adipocyte sizes. Lastly, patches showing excess blood were detected by identifying pixels with RGB values beyond the range expected of normal haematoxylin and eosin (H&E) stains.
Stain normalization
Potential stain variations between slides were reduced using the stain normalization methods from Macenko et al., which are commonly used in digital pathology32,33,34,35. In brief, this involved transforming images using predetermined stain vectors (Fig. 2).
Model architecture
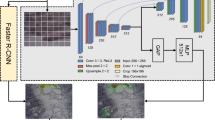
Our model employed a two-stage architecture comprising a CNN component for extracting discriminative features at the patch level, followed by a RNN component for aggregating patch-level features to produce an overall prediction for each whole-slide image (Fig. 3). By arranging the patches in a row-wise sequence, the RNN component can potentially learn the spatial arrangement of lesional patches within each slide. A similar architecture has been used for classifying histopathological images of gastric and colonic epithelial lesions24.
Convolutional neural network for feature extraction
The ResNet-50 architecture with an input size of 224 × 224 pixels was employed36. The ResNet-50 layers were then followed by the global average pooling layer, followed by two fully connected layers before terminating in two output nodes representing the FA and PT classes (Fig. 4A). Patch-level activation values from the global average pooling layer are intended to serve as representations of features learnt by the CNN model. The CNN model was first initialized with weights from the ImageNet database, and then further trained on the training data subset. The training subset was augmented with random vertical and horizontal image flips to make the model potentially more robust against variations in position and orientation.
A Architecture of convolutional neural network component. Numbers denote the dimensions of each layer. The global average pooling layer is shown in bold. B Row-wise arrangement of patch-level features that were fed into the recurrent neural network. Dashed lines denote valid patches generated from whole-slide image. C Architecture of recurrent neural network component. Numbers denote the dimensions of each layer. Patch activations were obtained from the global average pooling layer in the convolutional neural network component.
Recurrent neural network model for whole-slide classification
Activation values from the global average pooling layer in the CNN model (Fig. 4A) are taken as inputs to the RNN model. The RNN model consists of two Long Short-Term Memory (LSTM) layers with a hidden state size of 128 (Fig. 4C)37. LSTM was also used by Iizuka et al.24 for WSI analysis. During training, patches were arranged row-wise (Fig. 4B) and fed into the RNN model slide by slide with a batch size of one, i.e., each batch of training inputs comprised all patch activations for a given slide.
Model training and tuning
The CNN and RNN components of our model were trained sequentially. First, the CNN model was trained using patches with slide-level labels. A weighted cross entropy loss function was used to account for the imbalance in the relative scarcity of PT slides. Similar to the CNN model, a weighted cross entropy loss function was used to account for the class imbalance on the slide level. Both the CNN and RNN models were trained using the Adam optimization algorithm with momentum decay rates β1 = 0.9, β2 = 0.999 and an epsilon of 1 × 10−738. Both the CNN and RNN were trained for a maximum of 1000 epochs, with the possibility of early stopping when validation loss ceased to decline with additional training epochs.
Optimal model hyperparameters were determined empirically with the criterion of maximizing slide-level classification accuracy of the validation subset. We experimented with combinations of the following hyperparameters: batch size (16, 32), CNN learning rate (0.0001, 0.001) and RNN learning rate (0.0001, 0.001). Two metrics were used to examine the influence of hyperparameters on accuracy with respect to the testing subset. The importance metric characterizes non-linear relationships between hyperparameters and test accuracy while accounting for potential interactions between hyperparameters. We additionally examined the correlation metric, which quantifies linear relationships without accounting for interactions. Importance and correlation metrics were calculated based on implementations in the experiment tracking software Weights & Biases39.
Model evaluation
We evaluated patch- and slide-level predictions of our trained model on the testing subset by calculating the following metrics: per-class accuracy, true positive rates per class, and area under the receiver operating characteristic curve (AUC). Per-class accuracy refers to the percentage of correctly classified cases for the specified class of interest, i.e., FA or PT. In our calculations of true positive rates and AUC, we considered PT to be the positive class. Additionally, we regarded true positive (TP) as the number of PT cases predicted as PT; false negative (FN) as the number of PT cases predicted as FA; false positive (FP) as the number of FA cases predicted as PT; and true negative (TN) as the number of FA cases predicted as FA. The true positive rate (TPR) or recall (RE) was therefore defined as: TPR = TP/(TP + FN). Relatedly, the false positive rate (FPR) was defined as: FPR = FP/(FP + TN).
The receiver operating characteristic (ROC) curve plots the tradeoff between TPR and FPR at different probability thresholds for classification. As this threshold is lowered, more samples are classified as positive, thus increasing both true positive and false positive (i.e., FA cases predicted as PT), which in turn changes the TPR and FPR. The AUC is defined as the area under the ROC curve, with values ranging 0 to 1. An AUC value of 0.5 indicates classification performance equivalent to that of a 50:50 coin toss, while an AUC value of 1 denotes perfect classification performance.
Other metrics measured include precision and F1-score, defined as follows: Precision (PR) = TP/(TP + FP), and F1-score = 2 × ((PR × RE)/(PR + RE)).
Model performance
We performed another prediction method to compare with our main CNN and RNN approach. This was based on CNN and majority voting among the 224 × 224 pixels patches, in which if more than 50% of patches derived from a WSI are classified as PT (i.e., based on CNN on patches alone), then the whole slide is considered PT.
We also examined the speed of the model run on unannotated WSIs. The model was deployed as a minimum viable model (MVM) packaged into a Docker image and run on Docker Desktop for Windows version 3.4.0. The analysis was performed directly on WSIs scanned at 400× magnification in iSyntax file format downloaded from Philips PIPS. The machine used has Intel Core i9-9880H 2.30 GHz processor with 16GB RAM, running Windows 10 Pro Version 20H2. No dedicated GPU processing power was needed to run the algorithm. We ran the test set of 40 WSIs (file size ranging from about 240MB to 2.4GB) as a batch on the MVM in Docker.
Results
Our study cohort included WSIs of 187 FA biopsies, comprising 123 (66%) FA, 1 (1%) cellular FA, 61 (33%) FEL favoring FA, and 2 (1%) cellular FEL favoring FA (Table 1). As for the 100 PTs, there were 12 (12%) PT, 36 (36%) FEL favoring PT, 33 (33%) FEL, 8 (8%) cellular FEL favoring PT, and 11 (11%) cellular FEL (Table 2).
After applying patch filtering to exclude artifacts, and inclusion of areas within lesional annotations that demarcate discriminative regions, a total of 9228 FA patches and 6443 PT patches (each patch of 224 × 224 pixels) were extracted and generated from 133 FA and 64 PT WSIs respectively for the training subset. This represents a ~40-fold reduction compared to the number of patches potentially generated if filtering and annotations were not applied.
Our model tuning experiments showed that optimal results were obtained when the model was trained with a batch size of 32, CNN learning rate of 0.001 and RNN learning rate of 0.0001. The importance and correlation metrics both showed that batch size had the greatest association with test accuracy, followed by CNN learning rate and RNN learning rate (Supplementary Fig. 2). Batch size had a positive association with accuracy in terms of importance, but was negatively correlated with test accuracy, which suggests a non-linear relationship and/or interaction effects. When evaluated on the unannotated testing subset (20 FAs and 20 PTs), the trained CNN component had an overall patch-level accuracy of 68.1%, with 73.6% accuracy for FA patches and 65.1% accuracy for PT patches (Table 3). Test predictions had true positive rates of 81.2% for PT patches at the 50% classification threshold. When evaluating on the testing subset, the AUC for the CNN component was 0.693 (Fig. 5). Aggregating features extracted by the CNN component, the trained RNN component gave an overall slide-level accuracy of 87.5% on the unannotated testing subset, with accuracies of 80.0% and 95.0% for FA and PT slides, respectively (Table 3, Supplementary Tables 1 and 2). The model obtained a true positive rate/recall of 95% on PT slides at the 50% classification threshold, with 0.826 and 0.884 for its precision and F1-score respectively. The test AUC for the RNN component was 0.875 (Fig. 5). The 17 cases predicted as FA have a mean of 0.930 (95% CI 0.863–0.998) for their FA probability, and a mean of 0.070 (95% CI 0.002–0.137) for their PT probability. Meanwhile, 23 cases predicted as PT have a mean of 0.091 (95% CI 0.036–0.145) for their FA probability, and a mean of 0.909 (95% CI 0.855–0.964) for their PT probability. 3 FAs and 1 FEL favoring FA were misclassified as PT, while 1 benign PT was incorrectly predicted as FA. When the discordant slides were reviewed, their actual diagnoses remain the same, except the single PT case which was predicted as FA was now deemed to favor cellular FA. There were foci of epithelial tubule aggregation in areas boxed as PT (within FA cases) and inflamed stroma which may have contributed to the misclassification. One FA core biopsy had areas of mildly increased peri-epithelial stromal accentuation, but this would not raise concern for PT histologically. When the predicted results were based on CNN and majority voting method, the slide-level accuracy was lower at 80%, with TPR, precision and F1-score of 0.80 (Supplementary Table 2). Using this method similarly gave a discordant result for previously mentioned FAs, i.e., 3 FAs and 1 FEL favoring FA were misclassified as PT, while 1 PT, 2 FELs favoring PT and 1 cellular FEL favoring PT were predicted as FA. Our main CNN and LSTM method thus performed better.
Receiver operating characteristic curves (blue line) for patch-level predictions (A) and slide-level predictions (B) respectively. Phyllodes tumors were considered the positive class. Axes denote true positive rates and false positive rates along probability thresholds for FA and PT classification. The orange 1:1 line shows the baseline performance of a random, “coin toss” classifier.
The batch running time of the testing subset (n = 40) was 63 min, and each WSI took ~1 min 35 s to be processed, which is considered as a reasonable speed for clinical deployment.
Discussion
Making a distinction between cellular FAs and benign PTs in the preoperative setting still poses a challenge to breast radiologists, surgeons and pathologists, with the patient being the beneficiary of an enhanced diagnostic tool. A more objective and rapid detection tool could thus assist pathologists in accurately diagnosing the lesion in CNBs, without having to opt for excision. This may potentially bring significant cost savings, and reduces the need for surgical management and anxiety in patients. It is especially pertinent for women of reproductive age and adolescents who are more likely to develop FAs, compared to an older age group. This study focused on FAs and PTs on core biopsies, with the latter including some borderline PTs as well, since benign and borderline PTs share overlapping features which may not be possible to separate especially on limited core biopsy material, with final grading achieved only on excision specimens. Previous studies using digital pathology have evaluated mitosis-counting in PTs40, while digital point counting of stromal cellularity and expansion was not useful in the classification of FELs20. Application of artificial intelligence (AI) for FELs has so far involved analyzing ultrasound images via a deep learning software41, while Chan et al. developed an automatic support vector machine (SVM) algorithm for analyzing histopathology slides, with the use of fractal dimension to classify eight types of benign and malignant breast tumors27. A web-based BreaKHIS dataset of 7909 histopathological images (700 × 460 pixels PNG image) from 82 breast cancer patients was used. These comprised adenosis, FA, PT and tubular adenoma for benign tumors, while the malignant cohort included ductal carcinoma, lobular carcinoma, mucinous carcinoma and papillary carcinoma. Xie et al. similarly used this dataset, and performed deep learning techniques for supervised and unsupervised deep convolutional neural networks28. In both these studies, benign tumor subtypes were compared against malignant tumors and breast carcinomas. In contrast, Zheng et al. developed a deep learning classifier to distinguish between healthy breast tissue, non-neoplastic lesions, and 13 breast tumor subtypes, including benign FA and PTs (similar to the primary task in our study)29. Their network achieved 96.4% accuracy on the above 15-class classification task, with accuracies of 96.0% and 93.3% for FAs and PTs, respectively. These performance metrics were based on evaluation on an expert-annotated testing subset, and only sampled images extracted from annotated areas of WSIs were analyzed rather than directly on WSIs themselves; in contrast our model was evaluated on an unannotated testing subset of WSIs. The performance of our model may therefore more closely reflect clinical use-cases and processes in which time-consuming manual annotations are unfeasible. Another difference is that the Zheng et al. model was designed to specifically learn features from tightly-cropped patches centered on cell nuclei, while our model is potentially capable of extracting histological features in a more general manner. This is because our CNN model was built based on the highly generalizable ResNet 50 architecture, which has been successfully adapted for use across a diverse range of medical domains42,43,44. Furthermore, our model was trained on entire annotated lesional areas containing a variety of histological features as opposed to just nuclei. Together, these factors may possibly explain our model’s ability to attain comparable performance despite the use of an unannotated testing subset. In addition, the Zheng et al. model differs in using maximum pooling operations to aggregate features from patch to slide level. This involves summarizing patch activations by taking local maximum values. We hypothesize that this may overemphasize large activation values, and may not adequately account for dependencies between spatially distant patches. In contrast, our model uses LSTMs, which are able to more flexibly aggregate features and are known to be effective in relating information across longer (spatial) sequences37. Iizuka et al. employed a similar two-stage CNN-RNN model architecture for classifying gastric and colonic epithelial tumors24. Their model achieved an AUC of up to 0.97 and 0.99 for gastric adenocarcinoma and adenoma, respectively and 0.96 and 0.99 for colonic adenocarcinoma and adenoma respectively. The AUC for our model (0.875) for classifying cellular FAs and benign PTs is lower in comparison. The difference in performance could be attributed to the different types of data used. Iizuka et al. trained their model on a mix of surgical biopsy and resection data while our model is trained on only CNB data. The larger tissue size from inclusion of resection presents more information in each slide from which the model could more easily learn discriminative features for the classification task. In contrast, the smaller tissue size from CNB data could pose a challenge for the model to learn certain discriminative features such as the presence of leafy epithelium lined fronds which is a key diagnostic feature for PT1. In addition, the Iizuka et al. model was trained using a larger dataset (~4000 slides) which could have contributed to the model’s ability to generalize on unseen data. Furthermore, in discriminating between adenocarcinoma from adenoma, there is less likely to be an issue of lesional diagnostic feature representation compared to CNBs of breast FELs, the latter being compounded by intralesional heterogeneity where FAs and benign/borderline PTs may substantially show overlapping features between different areas of the same lesion. Given the rarity of PT, our study was limited by relatively fewer PT slides, thereby affecting the ratio of FA to PT. We applied class weights to manage this imbalance. Despite this, the strength of our work includes this being the first study which utilized AI to evaluate core biopsy images of FA and PT, while previous studies used datasets taken from partial mastectomy or excisional biopsy27,28,29. Performing annotations on more cases could possibly help to refine and improve the model, by supplying more training data in the future. Furthermore, evaluating samples from other institutions/laboratories would help us evaluate the applicability of the model beyond our institution, which would be considered for further study. Future studies may consider improving upon aspects of the AI model presented in this study. Firstly, some discriminative features differentiating FA and PT may require different magnification factors, e.g., increased stromal cellularity and atypia may be better observed at higher magnification factors while the presence of leafy fronds may require lower magnification. As the model developed in this study only uses a single magnification factor, it may not fully capture all discriminative features. A potential improvement would be to implement multiple CNN models with input images at different magnification factors during the feature extraction stage45. This could improve classification performance as the model will be able to better pick out discriminative features from the input images.
Secondly, the RNN architecture presented in this study was designed to only model unidirectional sequences of patch-level features along the horizontal axis of the whole-slide image. Future studies could investigate if bidirectional46 and/or two-dimensional LSTM models47 may potentially be more effective in modelling the spatial structure of patch-level features. Despite limitations, our study affirms the potential role of AI in facilitating diagnostic discrimination between FA and PT on core biopsy material which may be further refined for application in routine practice.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to institutional requirements governing data sharing, but are available from the corresponding author on reasonable request. Supplementary information is available at Laboratory Investigation’s website.
References
WHO Classification of Tumours Editorial Board. WHO classification of tumours of the breast, 5th edn. Lyon: IARC Press; 2019.
Tan, P. H. Fibroepithelial lesions revisited: implications for diagnosis and management. Mod. Pathol. 34, 15–37 (2021).
Tan, B. Y. et al. Phyllodes tumours of the breast: a consensus review. Histopathology 68, 5–21 (2016).
Jacklin, R. K. et al. Optimising preoperative diagnosis in phyllodes tumour of the breast. J. Clin. Pathol. 59, 454–459 (2006).
McCarthy, E. et al. Phyllodes tumours of the breast: radiological presentation, management and follow-up. Br. J. Radiol. 87, 20140239 (2014).
Yasir, S. et al. Cellular fibroepithelial lesions of the breast: a long term follow up study. Ann. Diagn. Pathol. 35, 85–91 (2018).
Komenaka, I. K., El-Tamer, M., Pile-Spellman, E. & Hibshoosh, H. Core needle biopsy as a diagnostic tool to differentiate phyllodes tumor from fibroadenoma. Arch. Surg. 138, 987–990 (2003).
Jacobs, T. W. et al. Fibroepithelial lesions with cellular stroma on breast core needle biopsy: are there predictors of outcome on surgical excision? Am. J. Clin. Pathol. 124, 342–354 (2005).
Dillon, M. F. et al. Needle core biopsy in the diagnosis of phyllodes neoplasm. Surgery 140, 779–784 (2006).
Lee, A. H., Hodi, Z., Ellis, I. O. & Elston, C. W. Histological features useful in the distinction of phyllodes tumor and fibroadenoma on needle core biopsy of the breast. Histopathology 51, 336–344 (2007).
Morgan, J. M., Douglas-Jones, A. G. & Gupta, S. K. Analysis of histological features in needle core biopsy of breast useful in preoperative distinction between fibroadenoma and phyllodes tumour. Histopathology 56, 489–500 (2010).
Jara-Lazaro, A. R. et al. Predictors of phyllodes tumours on core biopsy specimens of fibroepithelial neoplasms. Histopathology 57, 220–232 (2010).
Resetkova, E., Khazai, L., Albarracin, C. T. & Arribas, E. Clinical and radiologic data and core needle biopsy findings should dictate management of cellular fibroepithelial tumors of the breast. Breast J. 16, 573–580 (2010).
Ward, S. T. et al. The sensitivity of needle core biopsy in combination with other investigations for the diagnosis of phyllodes tumours of the breast. Int. J. Surg. 10, 527–531 (2012).
Gould, D. J. et al. Factors associated with phyllodes tumor of the breast after core needle biopsy identifies fibroepithelial neoplasm. J. Surg. Res. 178, 299–303 (2012).
Van Osdol, A. D. et al. Determining whether excision of all fibroepithelial lesions of the breast is needed to exclude phyllodes tumor: upgrade rate of fibroepithelial lesions of the breast to phyllodes tumor. JAMA Surg. 149, 1081–1085 (2014).
Tan, B. Y. et al. Morphologic and genetic heterogeneity in breast fibroepithelial lesions-a comprehensive mapping study. Mod. Pathol. 33, 1732–1745 (2020).
Pareja, F. et al. Phyllodes tumors with and without fibroadenoma-like areas display distinct genomic features and may evolve through distinct pathways. NPJ Breast Cancer 3, 40 (2017).
Lawton, T. J. et al. Interobserver variability by pathologists in the distinction between cellular fibroadenomas and phyllodes tumors. Int. J. Surg. Pathol. 22, 695–698 (2014).
Dessauvagie, B. F. et al. Interobserver variation in the diagnosis of fibroepithelial lesions of the breast: a multicentre audit by digital pathology. J. Clin. Pathol. 71, 672–679 (2018).
Tan, W. J. et al. A five-gene reverse transcription-PCR assay for pre-operative classification of breast fibroepithelial lesions. Breast Cancer Res. 18, 31 (2016).
Sim, Y. et al. A novel genomic panel as an adjunctive diagnostic tool for the characterization and profiling of breast Fibroepithelial lesions. BMC Med. Genom. 12, 142 (2019).
Ng, C. C. Y. et al. Genetic differences between benign phyllodes tumors and fibroadenomas revealed through targeted next generation sequencing. Mod. Pathol. 34, 1320–1332 (2021).
Iizuka, O. et al. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1–11 (2020).
Dimitriou, N., Arandjelović, O. & Caie, P. D. Deep learning for whole slide image analysis: an overview. Front. Med. 6, 264 (2019). Erratum in: Front Med (Lausanne) 7, 419 (2020).
Ianni, J. D. et al. Tailored for real-world: a whole slide image classification system validated on uncurated multi-site data emulating the prospective pathology workload. Sci. Rep. 10, 3217 (2020).
Chan, A. & Tuszynski, J. A. Automatic prediction of tumour malignancy in breast cancer with fractal dimension. R Soc. Open Sci. 3, 160558 (2016).
Xie, J., Liu, R., Luttrell, J. IV & Zhang, C. Deep learning based analysis of histopathological images of breast cancer. Front. Genet. 10, 80 (2019).
Zheng, Y. et al. Feature extraction from histopathological images based on nucleus-guided convolutional neural network for breast lesion classification. Pattern Recognit. 71, 14–25 (2017).
Han, Z. et al. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci. Rep. 7, 4172 (2017).
Bautista, P. A. & Yagi, Y. Improving the visualization and detection of tissue folds in whole slide images through colour enhancement. J. Pathol. Inform. 1, 25 (2010).
Macenko, M., et al. A method for normalizing histology slides for quantitative analysis. Proceedings of the IEEE International Symposium on Biomedical Imaging, 1107-1110 (IEEE, 2009).
Mahbod, A., Ellinger, I., Ecker, R., Örjan, S. Breast cancer histological image classification using fine-tuned deep network fusion. In: Campilho, A., Karray, F.& Romney, B. T. H., eds. Image analysis and recognition. p. 754–762 (Springer, Switzerland, 2018).
Rakhlin, A., Shvets, A., Iglovikov, V., Kalinin, A. A. Deep convolutional neural networks for breast cancer histology image analysis. In: Campilho, A., Karray, F. & Romney, B. T. H., eds. Image Analysis and Recognition. p. 737-744 (Springer, Switzerland, 2018).
Wang, Y., Dong, N., Dai, W., Rosario, S. D., Xing, E. P. Classification of breast cancer histopathological images using convolutional neural networks with hierarchical loss and global pooling. In: Campilho, A., Karray, F., Romney, B. T. H., eds. Image Analysis and Recognition. p. 845–852 (Springer, Switzerland, 2018).
He, K., Zhang, X., Ren, S., Sun, J. Deep residual learning for image recognition. In: Proceedings of the IEEE Computing Society Conference on Computer Vision and Pattern Recognition, 770–778 (CVPR, 2016).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Kingma D. P., Ba, J. Adam: A method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Biewald L. Experiment tracking with weights and biases. Weights and Biases, 2020. http://wandb.com/.
Chow, Z. L. et al. Counting mitoses with digital pathology in breast phyllodes tumors. Arch. Pathol. Lab. Med. 144, 1397–1400 (2020).
Stoffel, E. et al. Distinction between phyllodes tumor and fibroadenoma in breast ultrasound using deep learning image analysis. Eur. J. Radiol. Open 5, 165–170 (2018).
Reddy, S. B., Juliet, D. S. Transfer learning with ResNet-50 for malaria cell-image classification. In: IEEE International Conference on Communication and Signal Processing. 949 (IEEE, 2019).
Ferreira, C. A. et al. Classification of breast cancer histology images through transfer learning using a pre-trained inception Resnet V2. In: Campilho, A., Karray, F., & ter Haar Romeny, B., eds. Image Analysis and Recognition. ICIAR 2018. Lecture Notes in Computer Science, vol. 10882. p. 763–770. (Springer, Switzerland, 2018).
Hong, J., Cheng, H., Zhang, Y. D. & Liu, J. Detecting cerebral microbleeds with transfer learning. Mach. Vis. Appl. 30, 1123–1133 (2019).
Huang, W. C. et al. Automatic HCC detection using convolutional network with multi-magnification input images. In: 2019 IEEE International Conference on Artificial Intelligence Circuits and Systems (AICAS), 194-198 (IEEE, 2019).
Pratiher, S., Chattoraj, S., Agarwal, S., Bhattacharya, S. Grading tumor malignancy via deep bidirectional LSTM on graph manifold encoded histopathological image. In: IEEE International Conference on Data Mining Workshops, 674–681 (IEEE, 2018).
Kong, B., Wang, X., Li, Z., Song, Q., Zhang, S. Cancer metastasis detection via spatially structured deep network. In: Niethammer, M., Styner, M., Aylward, S., Zhu, H., Oguz, I., Yap, P. T. & Shen, D., eds. Information Processing in Medical Imaging. p. 236–248 (Springer, Switzerland, 2017).
Funding
This research / project is supported by the National Research Foundation, Singapore under its AI Singapore Programme (AISG Award no: AISG-100E-2020-053) and Singapore General Hospital (GB20CEOCF007). Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not reflect the views of National Research Foundation, Singapore.
Author information
Authors and Affiliations
Contributions
C.L.C., P.H.T., G.J.Z.N., S.R., K.H.N. and K.S.L.O. were responsible for study conceptualization and design, and project supervision. G.J.Z.N., K.W.J.C., Y.L., J.R., N.D.M.N., A.A.T., S.Y.H., V.C.Y.K., J.X.L., V.J.N.H., R.S., B.Y.T., and T.K.Y.T. performed the experiments. C.L.C., G.J.Z.N., K.W.J.C., Y.L., and J.R. analyzed and interpreted the data. C.L.C., N.D.M.N., G.J.Z.N., K.W.J.C., Y.L., and J.R. wrote the manuscript. All the authors revised and provided input to the manuscript.
Corresponding author
Ethics declarations
Competing interests
PHT holds patent applications for PCT/SG2015/050107 (Breast fibroadenoma susceptibility mutations and use thereof) and PCT/SG2015/050368 (Method and kit for pathologic grading of breast neoplasms). SGH collaborates with Royal Philips on a digital and computational pathology center of excellence and is a digital pathology reference site for Royal Philips. Other authors declare no competing interests.
Ethical approval
This study was approved by the SingHealth Centralized Institutional Review Board (CIRB Ref: 2019/2543), with a waiver of informed consent granted.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cheng, C.L., Md Nasir, N.D., Ng, G.J.Z. et al. Artificial intelligence modelling in differentiating core biopsies of fibroadenoma from phyllodes tumor. Lab Invest 102, 245–252 (2022). https://doi.org/10.1038/s41374-021-00689-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00689-0
This article is cited by
-
Discrimination between phyllodes tumor and fibro-adenoma: Does artificial intelligence-aided mammograms have an impact?
Egyptian Journal of Radiology and Nuclear Medicine (2022)