Abstract
Colorectal cancer (CRC) is the third leading cause of cancer-related death worldwide. Dysregulation of circular RNAs (circRNAs) appears to be a critical factor in CRC progression. However, mechanistic studies delineating the role of circRNAs in CRC remain limited. In this study, qRT-PCR and western blot assays were used to measure the expression of genes and proteins. Migration, invasion, proliferation, and apoptosis were examined by wound-healing, transwell, CCK-8, colony formation, and flow cytometry assays, respectively. Molecular interactions were validated by a dual-luciferase report system. A xenograft animal model was established to examine in vivo tumor growth and lung metastasis. Our data indicated that circN4BP2L2 expression was increased in CRC tissues and cell lines. Notably, inhibition of circN4BP2L2 effectively inhibited proliferation, migration, and invasion of LoVo cells, and inhibited tumor growth and metastasis in vivo, whereas the forced expression of circN4BP2L2 facilitated the proliferation, migration, and invasion of HT-29 cells. Mechanistic studies revealed that circN4BP2L2 acted as a molecular sponge of miR-340-5p to competitively promote CXCR4 expression. Furthermore, inhibition of miR-340-5p reversed the anti-cancer effects of circN4BP2L2 or CXCR4 silencing. Our data indicated an oncogenic role of circN4BP2L2 in CRC via regulation of the miR-340-5p/CXCR4 axis, which may be a promising biomarker and target for CRC treatment.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a major cause of the mortality in humans worldwide. CRC is a highly heterogenous disease and its development is closely associated with genetic and epigenetic alternations [1], as well as harmful lifestyle choices such as smoking [2], unhealthy diet [3], excessive alcohol consumption [4]. However, the underlying mechanisms for CRC development are still largely unknown. Therefore, it is imperative to investigate novel molecular mechanisms of CRC for the further development of new therapeutic strategies to delay disease progression.
Circular RNAs (circRNAs) are a type of non-coding RNA with a closed-loop structure and are extensively expressed in eukaryotic cells. Importantly, many circRNAs are abnormally expressed in CRC and to be involved in its initiation and progression. For example, Chen et al. showed that circ_001971 was increased in CRC and contributed to the growth, invasion, and angiogenesis of CRC in vitro and in vivo [5]. Wu et al. revealed that upregulation of circAPLP2 facilitated the growth and liver metastases of CRC cells via regulating miR-101-3p/Notch1 axis [6]. hsa_circ_0000471, as known as circN4BP2L2, is a newly identified circRNA originated from N4BP2L2 gene [7]. Li et al. have verified that circN4BP2L2 was aberrantly expressed in epithelial ovarian cancer and served as a prognostic biomarker [8]. However, the functional roles of circN4BP2L2 in CRC progression remain unclear.
MicroRNAs (miRNAs) are endogenous post-transcriptional regulators that mediate mRNA degradation or translation repression through binding to the 3ʹ untranslated region (UTR) of their target mRNAs [9]. In recent years, miRNAs have been widely implicated in CRC progression [10, 11]. For instance, Zhang et al. showed that miR-1258 was significantly downregulated in CRC tissues and cell lines, and overexpression of miR-1258 suppressed the proliferation and arrested cell cycle at G0/G1 phase by directly targeting the 3ʹ UTR of E2F8 [12]. Similarly, Li et al. illustrated that miR-875-3p was lowly expressed in CRC and correlated to higher rate of distant metastasis and worse prognosis, and ectopic expression of miR-875-3p attenuated the proliferative and migratory capacities of CRC cells by negatively controlling PLK1 levels [13]. By bioinformatic analysis, a potential interaction between miR-340-5p and circN4BP2L2 was predicted. However, the functional relevance of miR-340-5p and circN4BP2L2 remains to be further clarified.
It is interesting that circRNAs play an essential role in neutralizing cellular miRNAs by acting as competitive endogenous RNAs (ceRNAs), which therefore relieve the repressive effect of miRNAs on their genuine targets [14, 15]. Chemokine receptor 4 (CXCR4), a key chemokine receptor in cancer metastasis, was widely used in diagnosis and prognosis of CRC [16]. In the present study, the potential interaction between miR-340-5p and CXCR4 was predicted by Starbase database. Moreover, the regulatory network among circN4BP2L2, miR-340-5p, and CXCR4 was further explored. Our data demonstrated that circN4BP2L2 could enhance CXCR4 expression by absorbing miR-340-5p, and thereby promoting CRC cell proliferation and metastasis, which revealed an important role for circRNA-miRNA regulatory patterns in CRC tumorigenesis.
Material and methods
Clinical samples and cell culture
Human tumor tissues and para-cancerous tissues were collected from 32 CRC patients through surgical operation in the Xiangya Hospital, Central South University, which were frozen in liquid nitrogen immediately and stored at −80 °C for qRT-PCR detection. Written informed consent was signed by the patients before specimen collection. The study was approved by the Hospital’s Ethics Committee. The human colorectal cell lines (HT-29, SW480, HCT-116, and LoVo) and normal colon epithelial cells (NCM460) were commercially obtained from the Cell Bank of Chinese Academy of Science (Shanghai, China). All the cells were kept in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, USA) with 10% fetal bovine serum (FBS; Gibco, USA) and maintained in a CO2 incubator at 37 °C.
Vector construction and cell transfection
Human circN4BP2L2 cDNA was synthesized and inserted into the pLVX-cir vector (Genomeditech, Shanghai, China) to construct the circN4BP2L2-overexpressing plasmids. miR-340-5p mimics, miR-340-5p inhibitor, and short hairpin RNAs (shRNAs) targeting circN4BP2L2 or CXCR4 as well as their negative controls (mimics NC, inhibitor NC, sh-NC) used in this study were synthesized by GenePharma (Shanghai, China). For cell transfection, LoVo or HT-29 cells were transfected with the above plasmids or shRNAs using lipofectamine™ 2000 (Invitrogen, USA) following the manufacturer’s instructions. The transfection efficiency was evaluated by after transfection for 48 h.
Cell counting kit 8 (CCK-8) assay
Cell viability was determined using CCK-8 assay (Don Jindo) following the standard protocol. Briefly, 96-well plates were seeded with 1 × 103 cells in each well. After growth in CO2 incubator for 24, 48, 72 h, CCK-8 solution was added into each well, followed by incubation for another 2 h. Then, the absorbance at 490 nm was measured using a microplate reader (MRX; Dynex Technologies, West Sussex, UK).
Colony formation assay
Approximately 500 cells were seeded in a six-well plate in triplicate and cultured for 2 weeks. Cell cultivation was terminated until colonies could be observed under the inverted microscope. Thereafter, the colonies were fixed in 2 ml paraformaldehyde (4%) and stained with crystal violet (Sigma, USA). Eventually, the colonies in each well were counted.
Cell apoptosis assay
At 24 h after the transfection, CRC cells were harvested and washed with pre-cold PBS. The apoptotic cells were measured using the Annexin V FITC Assay Kit (Bio-Rad) according to the manufacturer’s instructions. Percentage of apoptotic cells was detected on a flow cytometer (BD Biosciences, USA).
Wound-healing assay
Wound-healing assay was performed as described previously [17]. Briefly, 5 × 105 cells were added into six-well plates and then cultured for 24–48 h. With 90% confluence, the medium was replaced to serum-free medium, and the cell monolayer was scratched with a sterile pipette tip. At the indicated time points (0, 24, and 48 h), the migration distance of cells was observed and captured under a light microscope (Olympus Corp., Tokyo, Japan).
Transwell assay
Transwell assay was performed to examine cell invasive ability using transwell chambers (BD Biosciences, San Jose, CA, USA). Generally, the upper chambers pre-coated with Matrigel were added with 200 μL serum-free DMEM medium containing 1 × 105 cells, and the lower chambers were added with 500 μL DMEM medium supplemented with 10% FBS. After 24 h, the non-invaded cells were removed with cotton swads, and the invaded cells in lower chambers were fixed with methanol and stained by crystal violet. Subsequently, ten fields were randomly chosen to count cell number under a microscope.
Quantitative real-time PCR (qRT-PCR) assay
RNAiso Plus (Takara Biotechnology, Co., Ltd) was used to extract total RNA from tissues and cells. One microgram of total RNA was reverse transcribed into cDNA using a cDNA synthesis kit (ThermoFisher). qRT-PCR was performed using the SYBR® Green system (Takara Biotechnology, Co., Ltd). The 2−ΔΔCt method was applied to calculate the relative expression levels of mRNA and miR-340-5p, using GAPDH and U6 RNA as reference genes, respectively. The primer sequences used in this experiment were listed as below:
circN4BP2L2 forward: 5ʹ-CAAAGACCTCCTCCTCCACA-3ʹ;
circN4BP2L2 reverse: 5ʹ-TCAGTGCTGAACACAATGCC-3ʹ;
miR-340-5p forward: 5ʹ-GCGGTTATAAAGCAATGAGA-3ʹ;
miR-340-5p reverse: 5ʹ-GTGCGTGTCGTGGAGTCG-3ʹ;
CXCR4 forward: 5ʹ-CCGTGGCAAACTGGTACTTT-3ʹ;
CXCR4 reverse: 5ʹ-GACGCCAACATAGACCACCT-3ʹ;
GAPDH forward: 5ʹ-CTGACTTCAACAGCGACACC-3ʹ;
GAPDH reverse: 5ʹ-GTGGTCCAGGGGTCTTACTC-3ʹ;
U6 forward: 5ʹ-CTGACTTCAACAGCGACACC-3ʹ;
U6 reverse: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Western blots
After washing with ice-cold PBS, the cells were lysed on ice in Tris Lysis Buffer (Cell Signaling, Danvers, MS) supplemented with protease inhibitor cocktail (Roche). After spinning down the debris, the supernatants were collected and quantified. Twenty micrograms of each protein sample was separated by 10% sodium dodecylsulphate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membrane. The membranes with transferred proteins were blocked in 5% bovine serum albumin buffer for 1 h, and then incubated with primary antibodies against CXCR4 (1:100, ab124824), cleaved caspase-3 (1:1000, ab49822), cleaved caspase-9 (1:1000, ab52298), E-cadherin (1:1000, ab40772), and Vimentin (1:1000, ab92547) overnight at 4 °C. After 3 times washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody. Finally, the bands of proteins were visualized by an ECL detection kit (Beyotime, China) and quantified using Image J software.
Dual-luciferase reporter gene assay
circN4BP2L2 and CXCR4 3′UTR containing miR-340-5p-binding sites were amplified using genomic DNA as template and cloned into the pmirGLO vector (circN4BP2L2-WT, CXCR4-WT) for luciferase reporter assay (Promega, Madison, WI). Mutation of circN4BP2L2 and CXCR4 sequences (circN4BP2L2-MUT, CXCR4-MUT) were amplified by PCR using synthetic primers. 293T cells were seeded in a 24-well plate, then transfected with the circN4BP2L2-WT, circN4BP2L2-MUT, CXCR4-WT, and CXCR4-MUT plasmids together with mimics NC or miR-340-5p mimics using Lipofectamine 2000 (Invitrogen). After transfection for 24 h, the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System Kit (Promega) in accordance with the provided instructions.
Xenograft animal models
BALB/c nude mice (4–5 weeks old, male) were purchased from Model Animal Research Center of Nanjing University (Nanjing, China). All mice were maintained on a pathogen-free animal facility under a 12 h light/12 h dark conditions. All experiments procedures in this study were approved by the Animal Care committee of Xiangya Hospital, Central South University. The mice were randomly divided into sh-NC and sh-CircN4BP2L2 groups (n = 6 per group). After anesthetization, sh-CircN4BP2L2-transfected LoVo cells or sh-NC-transfected LoVo cells (3 × 106) were subcutaneously injected into the back flanks of mice. Tumor volume was examined every 5 days for 1 month. After the injection for 1 month, the tumor tissues were obtained and weighed. For in vivo metastasis assay, sh-CircN4BP2L2-transfected LoVo cells or sh-NC-transfected LoVo cells (3 × 106) were injected into the tail veins of nude mice. After 5 weeks, mice were euthanized and lung tissues were collected to examine metastasis.
H&E staining assay
To evaluate metastasis, H&E staining was adopted. The lung tissues of mice were fixed in 4% paraformaldehyde, embedded into paraffin and cut into 4 μm. Then, a routine H&E staining was performed on the sections. The images were observed under a microscope.
Statistical analysis
One-way ANOVA was performed to analyze the statistical differences among multiple groups using GraphPad Prism (San Diego, CA). Student’s t test was used to assess the differences between two groups. All data were collected in triplicates, representing at least three independent experiments, and recorded as the mean ± SD. P values less than 0.05 were considered as statistically significant.
Results
circN4BP2L2 is upregulated in CRC tissues and cell lines
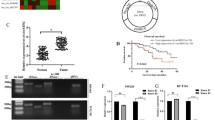
By circBank (http://www.circbank.cn/index.html) database analysis, hsa_circ_0000471 was spiced from host gene N4BP2L2 and with the Spliced length 388 bp. Recently, circN4BP2L2 was identified as a circRNAs that abnormal upregulated in CRC cells. Here, we firstly designed full-length amplification primers and to clone circN4BP2L2 gene and detected it by agarose gel electrophoresis (supplementary material, Fig. S1). Then, to explore the expression level of circN4BP2L2 in CRC, 32 pairs of CRC tissues and adjacent tissues were collected and analyzed by qRT-PCR. The results showed that circN4BP2L2 level in CRC samples was significantly higher than that in adjacent tissues (Fig. 1A). Furthermore, a higher circN4BP2L2 level was observed in stage III and IV CRC tissues as compared with that in stage I and II (Fig. 1B). As shown in Fig. 1C, circN4BP2L2 was upregulated in CRC tissues collected from patients with lymph node metastasis. In addition, the expression level of circN4BP2L2 in normal intestinal epithelial cell line (NCM460) and CRC cell lines (SW480, HT-29, LoVo, HCT-116) were further evaluated. Similarly, circN4BP2L2 was obviously upregulated in CRC cells as compared with normal NCM460 cells. Taken together, these results suggested that circN4BP2L2 was upregulated in CRC tissues and cell lines, and closely correlated with tumor stage and lymph node metastasis.
A circN4BP2L2 expression level in CRC tissues and paired non-tumor tissues was measured by qRT-PCR (n = 32). B qRT-PCR for determining circN4BP2L2 level in stage I, II, III, and IV CRC tissues. C circN4BP2L2 level was measured in CRC tissues of patients with or without lymph node metastasis. D The abundance of circN4BP2L2 in various CRC cell lines and normal intestinal epithelial cell line NCM460 was investigated using qRT-PCR. Data were collected from three independent experiments and arepresented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Silencing of circN4BP2L2 represses cell proliferation, migration and invasion, and enhances apoptosis of LoVo cells
To examine whether silencing of endogenous circN4BP2L2 could affect the biological functions of CRC cells, three shRNAs were designed to target the coding region of circN4BP2L2 (sh-circN4BP2L2). The knockdown efficiency was tested by qRT-PCR, of which sh-circN4BP2L2#1 was selected for further experiments due to its highest knockdown efficiency (Fig. 2A). CCK-8 and colony formation assays demonstrated that knockdown of circN4BP2L2 repressed the proliferation of LoVo cells (Figs. 2B, C). Flow cytometry assay showed that the population of apoptotic cells in LoVo cells transfected with sh-circN4BP2L2 was significantly increased compared with those transfected with sh-NC (Fig. 2D). Furthermore, wound-healing assay revealed that the migratory ability of LoVo cells was significantly suppressed by sh-circN4BP2L2 transfection (Fig. 2E). Similarly, transwell assay showed that depletion of circN4BP2L2 remarkably reduced the invasive ability of LoVo cells (Fig. 2F). These results indicate that depletion of endogenous circN4BP2L2 repressed cell proliferation, migration and invasion, while promoted apoptosis in LoVo cells.
A Of the three shRNAs, sh-circN4BP2L2#1 was identified as the most efficient one to knockdown circN4BP2L2 expression by qRT-PCR. B Growth curves were performed for circN4BP2L2 knockdown or control LoVo cells via CCK-8 assay. C Knockdown of circN4BP2L2 inhibited CRC cell proliferation, as shown by colony formation assay. D Attenuated circN4BP2L2 expression increased the population of apoptotic CRC cells through flow cytometry assay. E sh-circN4BP2L2 restrained migration of CRC cells by wound-healing assay. F A Transwell invasion assay was performed to suggest sh-circN4BP2L2 inhibited the invasion of LoVo cells. Data were collected from 3 independent experiments and presented as mean ± SD. *P < 0.05, **P < 0.01.
Overexpression of circN4BP2L2 promotes proliferation, migration, and invasion, but inhibits apoptosis in HT-29 cells
Here, we sought to investigate the effects of circN4BP2L2 overexpression on the malignant behaviors of HT-29 cells. As shown in Fig. 3A, cellular circN4BP2L2 expression level was enhanced by transfection with circN4BP2L2-overexpressing plasmids. Subsequently, cell proliferation assays suggested that overexpression of circN4BP2L2 markedly increased the proliferative capacity of HT-29 cells (Fig. 3B, C). Apoptotic assay demonstrated that circN4BP2L2 overexpression markedly reduced the percentage of apoptotic cells of HT-29 cells (Fig. 3D). We next assessed the migratory and invasive abilities of the circN4BP2L2-overexpressed cells using wound-healing and transwell assays. The results suggested a promoting effect of circN4BP2L2 on migration and invasion of HT-29 cells (Fig. 3E, F). These data indicated an important role of circN4BP2L2 in promoting HT-29 cell growth and metastasis.
A Overexpression of circN4BP2L2 in HT-29 cells was verified using qRT-PCR. B, C The proliferation of circN4BP2L2-expressing HT-29 cells was assessed by CCK-8 and colony formation assays. D The apoptosis of CRC cells after transfection with the circN4BP2L2 overexpression plasmid was detected by flow cytometry assay. E Cell migration was determined by a wound-healing assay. F A Transwell invasion assay was performed in HT-29 cells. Data were collected from three independent experiments and presented as mean ± SD. *P < 0.05, **P < 0.01.
Silencing of circN4BP2L suppresses CRC tumor growth and lung metastasis in vivo
Next, we examined the effect of circN4BP2L on a xenograft animal model. The results showed that knockdown of circN4BP2L resulted in a decrease of tumor volume and tumor weight compared to sh-NC group (Fig. 4A–C). qRT-PCR assay described that circN4BP2L in tumor tissues was significantly downregulated in sh-circN4BP2L group compared to sh-NC group (Fig. 4D). Moreover, LoVo cells treated differently were injected with a caudal vein to assess the effect of circN4BP2L on lung metastasis of tumor. As shown in Fig. 4E, F, compared to sh-NC group, knockdown of circN4BP2L significantly reduced the number of metastasis nodules in lung tissues of mice. Therefore, knockdown of circN4BP2L restrained CRC tumor growth and lung metastasis in vivo.
A Tumor image at 30 d after inoculation. B Tumor volume was calculated at the indicated time points. C Tumor weight at 30 d after inoculation. D Expression of circN4BP2L in tumor tissues was assessed by qRT-PCR. E Representative images of lung metastases of nude mice with tail vein injection. F Pathologic changes of lung metastasis of nude mice with tail vein injection were observed by H&E staining. Data were collected from three independent experiments and presented as mean ± SD. *P < 0.05, **P < 0.01.
circN4BP2L2 acts as a ceRNA by binding to miR-340-5p to regulate CXCR4 expression
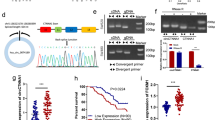
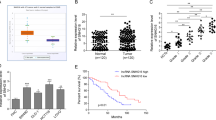
We next focused on the molecular mechanism underlying the biological functions of circN4BP2L2 in vitro. Intriguingly, we predicted a potential interaction between miR-340-5p and circN4BP2L2 by bioinformatic database (Starbase v2.0) (Fig. 5A). Luciferase assay showed that the miR-340-5p mimics strikingly inhibited the luciferase activity of circN4BP2L2-WT plasmid-transfected cells, however, no significant change was observed in circN4BP2L2-MUT-transfected cells (Fig. 5B). Moreover, we found a decreased abundance of miR-340-5p in CRC tissues as compared with that in adjacent tissues, and the decrease of miR-340-5p was associated to stage III/IV and lymph node metastasis (Fig. 5C, supplementary material, Fig. S2). Pearson correlation analysis suggested that circN4BP2L2 level was negatively correlated with miR-340-5p levels (Fig. 5D). As shown in Fig. 5E, we demonstrated that the relative expression of miR-340-5p was significantly enhanced in sh-circN4BP2L2 group. These data implied that circN4BP2L2 might absorb miR-340-5p. To explore the downstream targets of circN4BP2L2/miR-340-5p axis, starebase v2.0 database was employed. As described in Fig. 5F, the binding sites of miR-340-5p in 3′UTR region of CXCR4 were predicted. Furthermore, miR-340-5p mimics reduced the luciferase activity of cells transfected with CXCR4-WT plasmid, but not effects have found in cells transfected with CXCR4-MUT plasmids (Fig. 5G). Furthermore, qRT-PCR analysis showed that CXCR4 was upregulated in CRC tissues as compared with the adjacent tissues (Fig. 5H). Pearson correlation analysis suggested a negative correlation between miR-340-5p and CXCR4 (Fig. 5I). To further understand the role of circN4BP2L2/miR-340-5p axis in regulating CXCR4 expression, sh-circN4BP2L2 and miR-340-5p inhibitor were co-transfected into LoVo cells. As indicated in Fig. 5J, K, CXCR4 mRNA expression was pronouncedly decreased by transfection with sh-circN4BP2L2. Strikingly, the inhibitory effect of sh-circN4BP2L2 could be effectively rescued by miR-340-5p inhibitor. Collectively, these results suggested that circN4BP2L2 acted as a ceRNA by neutralizing miR-340-5p and thereby regulating CXCR4 expression in CRC cells.
A Predicted binding sites of miR-340-5p in circN4BP2L2 transcript. B miR-340-5p mimics or mimics NC was co-transfected into 293T cells together with the luciferase reporter vector. The relative luciferase activity was measured at 24 h post transfection. C qRT-PCR for detecting miR-340-5p levels in CRC tissues and paired non-tumor tissues. D The correlation between circN4BP2L2 and miR-340-5p was assessed by Pearson correlation analysis. E sh-circN4BP2L2 was transfected into CRC cells, and the expression of miR-340-5p was determined by qRT-PCR. F Putative miR-340-5p binding sites in the 3′ UTR of CXCR4. (G) A luciferase reporter vector encoding wild-type or mutaant type 3′ UTR of CXCR4 was co-transfected into 293 T cells together with miR-340-5p mimics or a negative control, and the relative luciferase activity was measured 24 h later. (H) CXCR4 levels in CRC tissues and paired non-tumor tissues was evaluated by qRT-PCR. I The correlation between CXCR4 and miR-340-5p was determined by Pearson correlation analysis. qRT-PCR (J) and western blot (K) for CXCR4 level in CRC cells transfected with sh-circN4BP2L2 or miR-340-5p inhibitor. Data were collected from three independent experiments and presented as mean ± SD. *P < 0.05, **P < 0.01.
circN4BP2L2 promotes CRC cell growth and metastasis through regulating miR-340-5p/CXCR4 axis
To investigate the functional relevance among circN4BP2L2, miR-340-5p, and CXCR4 in vitro, a series of rescue experiments were conducted. Silencing of endogenous circN4BP2L2 or CXCR4 significantly suppressed the proliferation and invasion of LoVo cells, which could be restored by miR-340-5p inhibitor (Fig. 6A, B). Notably, the proliferative and invasive abilities of CRC cells were pronouncedly enhanced by knocking down miR-340-5p alone (Fig. 6A, B). Western blot assay indicated that circN4BP2L2 or CXCR4 knockdown significantly increased cleaved caspase-3, cleaved caspase-9, E-cadherin expression, but decreased Vimentin expression in CRC cells, however, all of these changes were partly reversed by miR-340-5p inhibitor (Fig. 6C). Furthermore, we further verified the functions of circN4BP2L2/miR-340-5p/CXCR4 axis in HT-29 cells. As shown in Fig. 7A, B, overexpression of circN4BP2L2 or CXCR4 facilitated the proliferation and invasion of HT-29 cells, which were partly reversed by upregulation of miR-340-5p. The proliferative and invasive capacities of HT-29 cells was inhibited by miR-340-5p mimics. Protein levels of cleaved caspase-3, cleaved caspase-9, E-cadherin were enhanced, while vimentin levels were reduced in circN4BP2L2 or CXCR4-overexpressing HT-29 cells, and these effects were partly counteracted by miR-340-5p mimics (Fig. 7C). Collectively, these results indicate that circN4BP2L2 contributed to the growth and metastasis of CRC cells via regulating miR-340-5p/CXCR4 axis.
LoVo cells were transfected with sh-NC, sh-circN4BP2L2, sh-CXCR4, or in combination with miR-340-5p inhibitor. A The growth of transfected CRC cells was detected by colony formation assay. B The invasion of CRC cells from each group was assessed by a Transwell assay. C The protein levels of cleaved caspase-3, cleaved caspase-9, E-cadherin and Vimentin in CRC cells were assessed by western blotting. Data were collected from 3 independent experiments and presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
HT-29 cells were transfected with vector, circN4BP2L2, CXCR4, or in combination with miR-340-5p mimics. A The growth of CRC cells with various transfections was detected by colony formation assay. B The invasion of CRC cells from different groups was assessed by a Transwell assay. C The protein levels of cleaved caspase-3, cleaved caspase-9, E-cadherin and Vimentin in CRC cells were assessed by western blotting. Data were collected from three independent experiments and presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
CRC is a common malignancy with a high risk of recurrence and metastasis in humans [18]. At present, the CRC patients still have a poor prognosis due to the toxicity and limited impact on cure rates of current treatments [18]. Therefore, identification of new efficient molecular targets for the diagnosis and treatment of CRC are needed. Recent studies show that circRNAs are involved in the development and progression of CRC [19]. Chen et al. reported that circCTNNA1 accelerated the proliferation and invasion of CRC cells via regulating miR-149-5p/FOXM1 axis [20]. However, there is a limited understanding of the expression and functions of circRNAs in CRC. Metastasis is the major cause of death for CRC patients. During the process of metastasis, epithelial-mesenchymal transition (EMT) exerts crucial roles in tumor development [21]. Briefly, EMT refers to a transition from an epithelial morphology to a motile mesenchymal (migratory and invasive) phenotype, then facilitates cancer cells to permeate the basal lamina barrier and invade the adjacent tissue, which is related to downregulated epithelial cell marker (E-cadherin) and upregulated mesenchymal marker (vimentin) [22]. In this study, we focused on a novel circRNA, known as circN4BP2L2, which has been verified as a prognostic biomarker for ovarian cancer [8]. We demonstrated that circN4BP2L2 was upregulated in CRC tissues and cell lines and closely correlated with advanced TNM stage and lymph node metastasis. Further functional results suggested that circN4BP2L2 remarkably facilitated the growth and metastasis of CRC in vitro and in vivo. These findings indicated that circN4BP2L2 participated in the progression of CRC.
According to the ceRNA theory, circRNAs function as sponges for miRNAs at the transcriptional level, resulting in the increased expression of miRNA-targeted genes [23]. The mechanism of circRNA has been confirmed by a large number of studies. For example, a recent study documented that circ_101141 promoted hepatocellular carcinoma development via binding to miR-1297 to enhance ROCK1 expression [24]. Yin et al. revealed that circ-0046263 functioned as a ceRNA to absorb miR-133a-5p and increased IGFBP3 expression, which exerted tumor-promoting role in nasopharyngeal carcinoma [25]. The implication of miR-340-5p in tumor development and metastasis has been widely reported [26,27,28], including CRC [29]. In our study, miR-340-5p was downregulated in CRC tissues, and negatively related to stage III/IV and lymphatic metastasis, which was consistent to a study by Yang et al. [30]. In addition, we also identified that miR-340-5p expression in CRC tissue samples was negatively correlated with circN4BP2L2. Furthermore, bioinformatics analyses indicated that there were binding sites of miR-340-5p in circN4BP2L2 transcript, which was further experimentally verified by luciferase report assay. Moreover, silencing of circN4BP2L2 increased the expression of miR-340-5p. Rescue experiments further showed that the carcinogenic effects of circN4BP2L2 could be counteracted by miR-340-5p mimics. These results verified our speculation that circN4BP2L2 acts as a molecular sponge of miR-340-5p to affect the malignant behaviors of CRC cells.
Generally, miRNAs exert the biological functions through repressing translation or triggering the degradation of their target mRNAs. Intriguingly, we observed that CXCR4 was highly expressed in CRC tissues as compared with para-cancerous tissues, and negatively correlated with miR-340-5p expression. Notably, CXCR4 is a critical chemokine receptor responsible for cancer cell proliferation and metastasis [31, 32]. During CRC progression, several miRNAs such as miR-126 and miR-622 are reported to inhibit tumor metastasis by binding to CXCR4 3′UTR, indicating an important role of miRNA-CXCR4 regulatory pattern in CRC progression [33, 34]. In this study, CXCR4 was identified as a potential target of miR-340-5p by bioinformatics analysis, which was further confirmed by luciferase report assay. In addition, we also found that CXCR4 was a competitive target for circN4BP2L2 binding to miR-340-5p. Importantly, the depletion of CXCR4 impaired the proliferation and invasion of CRC cells, and CXCR4 negatively affected the biological roles of miR-340-5p in CRC cells. Collectively, these findings suggested that circN4BP2L2 facilitated CXCR4-mediated growth and metastasis by absorbing miR-340-5p in CRC.
In summary, our study determined that circN4BP2L2 functioned as an oncogene in CRC and correlates with lymphatic metastasis and advanced TNM stage. In addition, circN4BP2L2 promoted the growth and metastasis of CRC via regulating the miR-340-5p/CXCR4 axis (Fig. 8). Our study provided insight into the promising roles of circN4BP2L2 in CRC, which uncoverd a novel mechanistic elucidation of the ceRNA regulatory network in CRC progression and identifies a potential therapeutic target for CRC treatment.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Taylor DP, Burt RW, Williams MS, Haug PJ, Cannon-Albright LA. Population-based family history-specific risks for colorectal cancer: a constellation approach. Gastroenterology. 2010;138:877–85.
Liang PS, Chen TY, Giovannucci E. Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis. Int J Cancer. 2009;124:2406–15.
Chan DS, Lau R, Aune D, Vieira R, Greenwood DC, Kampman E, et al. Red and processed meat and colorectal cancer incidence: meta-analysis of prospective studies. PLoS ONE. 2011;6:e20456.
Fedirko V, Tramacere I, Bagnardi V, Rota M, Scotti L, Islami F, et al. Alcohol drinking and colorectal cancer risk: an overall and dose-response meta-analysis of published studies. Ann Oncol. 2011;22:1958–72.
Chen C, Huang Z, Mo X, Song Y, Li X, Zhang M. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res. 2020;39:91.
Wu HB, Huang SS, Lu CG, Tian SD, Chen M. CircAPLP2 regulates the proliferation and metastasis of colorectal cancer by targeting miR-101-3p to activate the Notch signalling pathway. Am J Transl Res. 2020;12:2554–69.
Zhong S, Zhou S, Yang S, Yu X, Xu H, Wang J, et al. Identification of internal control genes for circular RNAs. Biotechnol Lett. 2019;41:1111–9.
Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53:2637–46.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Kong F, Zou H, Liu X, He J, Zheng Y, Xiong L, et al. miR-7112-3p targets PERK to regulate the endoplasmic reticulum stress pathway and apoptosis induced by photodynamic therapy in colorectal cancer CX-1 cells. Photodiagnosis Photodyn Ther. 2020;29:101663.
Ou C, Sun Z, Li X, Ren W, Qin Z, Zhang X, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63.
Zhang Z, Li J, Huang Y, Peng W, Qian W, Gu J, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif. 2018;51:e12505.
Li SS, Zhu HJ, Li JY, Tian LM, Lv DM. MiRNA-875-3p alleviates the progression of colorectal cancer via negatively regulating PLK1 level. Eur Rev Med Pharmacol Sci. 2020;24:1126–33.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol Cancer. 2020;19:117.
Li LN, Jiang KT, Tan P, Wang AH, Kong QY, Wang CY, et al. Prognosis and clinicopathology of CXCR4 in colorectal cancer patients: a meta-analysis. Asian Pac J Cancer Prev. 2015;16:4077–80.
Ke B, Wang XN, Liu N, Li B, Wang XJ, Zhang RP, et al. Sonic Hedgehog/Gli1 signaling pathway regulates cell migration and invasion via induction of epithelial-to-mesenchymal transition in gastric cancer. J Cancer. 2020;11:3932–43.
Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065.
Bhuyan R, Bagchi A. Prediction of the differentially expressed circRNAs to decipher their roles in the onset of human colorectal cancers. Gene. 2020;762:145035.
Chen P, Yao Y, Yang N, Gong L, Kong Y, Wu A. Circular RNA circCTNNA1 promotes colorectal cancer progression by sponging miR-149-5p and regulating FOXM1 expression. Cell Death Dis. 2020;11:557.
Montserrat N, Mozos A, Llobet D, Dolcet X, Pons C, de Herreros AG, et al. Epithelial to mesenchymal transition in early stage endometrioid endometrial carcinoma. Hum Pathol. 2012;43:632–43.
Christofori G. New signals from the invasive front. Nature. 2006;441:444–50.
Lin Z, Long F, Zhao M, Zhang X, Yang M. The role of circular RNAs in hematological malignancies. Genomics. 2020;112:4000–8.
Zhang T, Zhang L, Han D, Tursun K, Lu X. Circular RNA hsa_Circ_101141 as a competing endogenous RNA facilitates tumorigenesis of hepatocellular carcinoma by regulating miR-1297/ROCK1 pathway. Cell Transplant. 2020;29:963689720948016.
Yin L, Chen J, Ma C, Pei S, Du M, Zhang Y, et al. Hsa_circ_0046263 functions as a ceRNA to promote nasopharyngeal carcinoma progression by upregulating IGFBP3. Cell Death Dis. 2020;11:562.
Liu Y, Li X, Zhang Y, Wang H, Rong X, Peng J, et al. An miR-340-5p-macrophage feedback loop modulates the progression and tumor microenvironment of glioblastoma multiforme. Oncogene. 2019;38:7399–415.
Kim S, Choi JY, Seok HJ, Park MJ, Chung HY, Bae IH. miR-340-5p suppresses aggressiveness in glioblastoma multiforme by targeting Bcl-w and Sox2. Mol Ther Nucleic Acids. 2019;17:245–55.
Lu G, Zhang Y. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell Mol Biol Lett. 2019;24:34.
Cheng B, Rong A, Zhou Q, Li W. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J Exp Clin Cancer Res. 2020;39:5.
Yang L, Men WL, Yan KM, Ti J, Nie YZ, Xiao HJ. MiR-340-5p is a potential prognostic indicator of colorectal cancer and modulates ANXA3. Eur Rev Med Pharmacol Sci. 2018;22:4837–45.
Yin X, Li Z, Zhu P, Wang Y, Ren Q, Chen H, et al. CXCL12/CXCR4 promotes proliferation, migration, and invasion of adamantinomatous craniopharyngiomas via PI3K/AKT signal pathway. J Cell Biochem. 2019;120:9724–36.
Zhu Y, Tang L, Zhao S, Sun B, Cheng L, Tang Y, et al. CXCR4-mediated osteosarcoma growth and pulmonary metastasis is suppressed by MicroRNA-613. Cancer Sci. 2018;109:2412–22.
Liu Y, Zho Y, Feng X, An P, Quan X, Wang H, et al. MicroRNA-126 functions as a tumor suppressor in colorectal cancer cells by targeting CXCR4 via the AKT and ERK1/2 signaling pathways. Int J Oncol. 2014;44:203–10.
Fang Y, Sun B, Wang J, Wang Y. miR-622 inhibits angiogenesis by suppressing the CXCR4-VEGFA axis in colorectal cancer. Gene. 2019;699:37–42.
Author information
Authors and Affiliations
Contributions
KY and YW performed the experiments and wrote the manuscript. FZ collected and analyzed the data. ZZ designed and supervised the study. BL contributed the methodology. DF and ZZ analyzed the data and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics statement/consent to participate
The study has been approved by the Hospital’s Ethics Committee. All patients were informed of the study and signed the written consent.
Funding statement
This work was supported by the Natural Science Foundation of Hunan Province (No. 2019JJ40480).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, KD., Wang, Y., Zhang, F. et al. CircN4BP2L2 promotes colorectal cancer growth and metastasis through regulation of the miR-340-5p/CXCR4 axis. Lab Invest 102, 38–47 (2022). https://doi.org/10.1038/s41374-021-00632-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00632-3
This article is cited by
-
CircRNAs in colorectal cancer: potential biomarkers and therapeutic targets
Cell Death & Disease (2023)
-
Circular RNA circ_0005835 promotes promoted neural stem cells proliferation and differentiate to neuron and inhibits inflammatory cytokines levels through miR-576-3p in Alzheimer’s disease
Environmental Science and Pollution Research (2022)
-
miR-340-5p mediates the therapeutic effect of mesenchymal stem cells on corneal neovascularization
Graefe's Archive for Clinical and Experimental Ophthalmology (2022)