Abstract
New cancer characteristics can be discovered by focusing on the process of tumor formation. Cancer stem cells (CSCs) are a key subpopulation, as they are theorized to be at the apex of the tumor hierarchy. We can better understand their function in the tumor hierarchy by using sectioned samples to observe the growth of tumors from their origins as CSCs. In this study, we evaluated the growth of moderate differentiated colorectal cancer from LGR5-positive cells, which is a CSC marker of colorectal cancer, using xenograft and three-dimensional culture models spatiotemporally. These cells express LGR5 at high levels and show CSC phenotypes. To detect them, we used a previously generated antibody that specifically targets LGR5, and were therefore able to observe LGR5-positive cells aggregating into small clusters (sCLs) over the course of tumor growth. Because these LGR5-expressing sCLs formed continuously during growth mainly in the invasive front, we concluded that the structure must contribute significantly to the expansion of CSCs and to tumor growth overall. We confirmed the formation of sCLs from gland structures using a three-dimensional culture model. In addition, sCLs exhibited upregulated genes related to stress response and partial/hybrid epithelial–mesenchymal transition (EMT), as well as genes reported to be prognosis factors. Finally, sCLs with high LGR5 expression were identified in clinical samples. Based on these results, we elucidate how sCLs are an important contributors to tumor growth and the expansion of CSCs.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide and has a high mortality rate [1]. As a subpopulation of tumor cells, cancer stem cells (CSCs) have been a major focus of research for over a decade [2,3,4]. In CSC theory, CSCs are the apex of the hierarchical structure can both maintain themselves through self-renewal and differentiate into non-CSCs, and are correlated with invasion/metastasis, drug/radio-resistance, and recurrence.
LGR5 is a known CSC marker [5]. It is thought to be potentiator of Wnt/β-catenin signaling [6] and its expression is also markedly increased in subsets of colorectal, liver, pancreatic, stomach, and endometrial cancers [7, 8]. Hence, LGR5 is considered a driving force in a subset of human cancers. On the other hand, although its role at specific stages in the progression of cancer is hard to dispute, its clinical relevance is still controversial [9]. One reason for this is that there has long been no antibody to target LGR5 [10], making it difficult to detect on sectioned slides.
Previously, we generated specific antibodies against human LGR5, successfully identifying one that could be used to detect LGR5 on clinical human samples [11, 12]. Furthermore, we established a human colon cancer cell line highly expressing LGR5 from a patient-derived xenograft (PDX) model using an adherent culture, and confirmed the CSC phenotype [11]. We think these research tools are very useful for understanding how tumors grow from CSCs and can also provide information about their distribution. This can deepen our understanding of the role of CSCs in tumor growth.
In CRC, adenocarcinoma is characterized by glandular formation, a typical histopathological structure [13]. Recently, unique structures such as tumor budding (TB) and poorly differentiated clusters (PDCs) have been investigated as independent prognosis factors [14,15,16,17]. TBs are defined as tumor cell clusters with fewer than five cells, and PDCs as consisting of five or more tumor cells without glandular structures. The relationship between TB/PDC and CSC or epithelial–mesenchymal transition (EMT) is generally accepted, though it is still under discussion [18,19,20,21].
In this study, we focused on the relationship between tumor structures, including TB/PDC and the distribution of LGR5, in the tumor growth process. In general, a structure’s appearance can differ greatly depending on the slice depth when diagnosis uses a single section [22, 23]. We evaluated tumor growth in a xenograft model derived from an LGR5-positive human colon cancer cell line using spatiotemporal observation.
Materials and methods
Cell lines
A stable human colon cancer cell line that expresses LGR5 and possesses CSC properties (PLR123) was cultured as previously described [11]. The LGR5-positive CSC line (PLR123) was established from a colon cancer PDX model after serial passages in NOD/Shi-scid, IL-2RγKO (NOG) mice and the subsequent adherent cell culture of these tumors. In addition, the PLR123 cell line was derived from moderately differentiated colon cancer. Human CRC cell line LS174T (ATCC CRL-188, VA, USA) and colon fibroblast CCD-18Co (ATCC CRL-1459) were cultured in minimum essential medium supplemented with 10% fetal bovine serum.
Growth process of xenotransplanted tumor tissues from LGR5-positive CSC line
Twenty-six NOG mice (CLEA Japan, Tokyo, Japan) were used in the experiment. To obtain xenograft models, 1 × 105 (day 3, 7, 12, and 19) or 1 × 103 (day 35) of PLR123 cells suspended in Hank’s balanced salt solution (Life technologies, MA, USA) with 50% Matrigel were subcutaneously inoculated into the flanks of mice. Mice were sacrificed by exsanguination under isoflurane anesthesia at 3 (n = 7), 7 (n = 7), 12 (n = 6), 19 (n = 3), and 35 (n = 3) days after the inoculation of PLR123 cells. At necropsy, the xenotransplanted tumor tissues were removed from each mouse. All animal experiments were approved by the Ethical Committee for Treatment of Laboratory Animals at Chugai Pharmaceutical Co., Ltd. (day 3, 7, 12, and 19) or Forerunner Pharma Research Co., Ltd (day 35).
Tissue preparation
Xenograft samples were divided along the largest cross-section, and fixed with 4% paraformaldehyde (PFA) at 4 °C for 16–24 h. All samples were processed and embedded in paraffin by the AMeX method [24, 25], and hematoxylin and eosin (HE)-stained sections were produced by the usual method. Serial sections (4 μm each) stained by HE were created for one sample taken at day 3 (55 slides), 7 (100 slides), and 12 (100 slides), and 3D analysis was performed. Another sample at day 12 was sliced into 100 serial sections and used for the immunofluorescence (IF) study (LGR5 and HLA-DMA). Furthermore, one sample in day 19 was sliced into 20 serial sections and used for IF (LGR5 and HLA-DMA) to analyze the 3D distribution of LGR5-positive cells.
The paraffin block derived from a study of the orthotopical inoculation into the cecum of NOG mice was provided by SK (Forerunner Pharma Research). In that study, the same cell line used in the current study (PLR123) was implanted into the cecum of mice. Tumor mass was sampled more than 100 days after inoculation.
Clinical samples from patients
Primary CRC paraffin blocks were obtained from the archive of the Department of Surgery and Science, Kyushu University Hospital, Fukuoka, Japan. The samples were collected from patients who had undergone surgery without chemotherapy during the period of 1998–2013 (Supplementary Table 1). The specimens were embedded into paraffin blocks after fixation in 10% neutral buffered formalin. This study was approved by the Institutional Review Board of Kyushu University (approval no. 27-280).
Detection of LGR5 on sectioned slides
It is well known that a very low level of LGR5 protein is expressed in tissues. Therefore, the following enhanced methods were performed to detect LGR5 as previously described [11, 12].
Briefly, after incubation with the primary antibody (2U2E-2, in-house, 2.5 μg/mL), the sections were incubated with a secondary antibody conjugated with biotin (Abcam, Cambridge, UK), and the reaction was visualized by Streptavidin-Qdot 605 (Thermo Fisher Scientific, MA, USA); or after incubation with the primary antibody (2U2E-2, 1.0 μg/mL), the sections were incubated with a secondary antibody conjugated with polymer-horseradish peroxidase (DAKO, CA, USA) and the reaction was visualized by Alexa Fluor 488-labeled tyramide (Thermo Fisher Scientific). All the specimens were also stained with Qnuclear deep red stain (Thermo Fisher Scientific) or DAPI to detect nuclei.
The H-score of LGR5 expression was calculated for clinical samples. We randomly selected four or five fields per slide stained for LGR5 expression for each patient sample. The intensity and the area of expression in each selected field were read by certified pathologists. Intensity was classified as weak, moderate, or strong, and the area for each intensity grade was estimated as a percentage of the whole field. Then, the H-score was calculated for each field as follows: H-score (0–300) = 0 × (% cells with negative expression) + 1 × (% cells with weak staining) + 2 × (% cells with moderate staining) + 3 × (% cells with strong staining). Finally, the H-score for each individual patient was calculated as the average H-score of four or five fields.
Immunofluorescence
Sectioned specimens from the above-mentioned paraffin blocks were incubated with anti-HLA-DMA antibody (Sigma-Aldrich, MO, USA), anti-E-cadherin (GeneTex, CA, USA), anti-β-catenin antibody (BD bioscience, CA, USA), anti-Ki67 antibody (Abcam), and anti-S100A4 antibody (Novus Biologicals, CO, USA). After incubation with the primary antibodies, sections were incubated with a secondary antibody conjugated with biotin (Vector Laboratories, CA, USA) and the proteins were visualized by Alexa Fluor 568-labeled streptavidin (Life technologies). Those specimens were also stained with DAPI.
Counting number of tumor cells in one section
HE-stained single section slides on day 3, 7, and 12 (n = 3) were scanned into an Aperio Scan Scope (Leica Biosystems, Wetzlar, Germany). The number of tumor cell nuclei in pathologist-selected tumor areas was counted automatically by the IHC Nuclear Algorithm in Aperio (Leica Biosystems), and the approximate curve was calculated by Microsoft Excel.
Counting number of LGR5-positive cells in one section
Photos were taken for 12 random fields on a single section slide stained with LGR5 and HLA-DMA at day 3, 7, and 12 (n = 3) by confocal microscope (C1, Nikon, Tokyo, Japan). The number of LGR5-positive and -negative cells was counted on printed sheets.
Analysis of microscopic three-dimensional structure of tumor using serial sections
Each field was defined using photos of HE-stained serial sections above mentioned (n = 1, day 3, 7, and 12). An ID number was assigned for each microscopic 3D structure for identification on printed sheets. If the lumen could not be discerned in the structure, only the external diameter was measured. But if the lumen was recognized, both external and internal diameter was measured. Furthermore, the structures and their types were counted as described below.
Each field was defined using 100 individual photos of LGR5 and HLA-DMA-stained serial sections (n = 1, day 12). More than ten small clusters (sCLs) with/without lumens were identified and their external and/or internal diameters were measured. Furthermore, the LGR5-positive rate was measured for each slide and translated into points [1–10%: 1, 11–35%: 2, 36–65%: 3, 66–95%: 4, >96%: 5], the average of which was expressed as a grade [rare: <1, occasional: 1 ≦ 2.5, moderate: 2.5 ≦ 4, frequent: ≧4]. Afterward, the cover glass was removed and stained by HE. Tumor morphology was then confirmed using a microscope.
Histopathological classification of tumor structures
To define structure type using a single slide, tumor cells composed on single slide were classified as sCLs or glands according to their pathological features [13, 26, 27]. Namely, an sCL consists of a few cell-aggregating structures with or without a lumen. And a gland has a lumen with an approximate diameter of over 100 μm, along with a regular epithelial cell arrangement and nuclei in the basal side of cytoplasm.
To define structures for 3D analysis using serial sections, structures were classified by size and morphological feature. A small aggregation of cells consisting of round/oval cells with or without a lumen with an approximate diameter of less than 100 μm is an “sCL.” A structure consisting of columnar cells and a lumen with an approximate diameter of more than 100 μm is a “gland.”
LGR5 signal intensity profile
The signal profiling function in NIS-element (Nikon) was used to measure the LGR5 signal profile and distance from the tumor interface to the center (0–500 μm) for samples taken on day 35 (n = 3). The signals were measured to be ~0.62 μm and the total of measured values was 100 μm.
Laser microdissection (LMD) and DNA microarrays
sCL-like structures (n = 3) and glands (n = 3) in the xenograft tissue were isolated by LMD (LMD7000, Leica Microsystems) on sectioned slides 12 days after subcutaneous inoculation of PLR123. The xenograft samples were sectioned at room temperature, floated in an RNase-free water bath, and transferred to frame slides (Leica Microsystems). After being stained by HE, sCL-like structures and glands were dissected by the LMD system. sCL-like structures and glands were distinguished by a pathologist. After extracting total RNA, biotin-labeled cRNA was produced from total RNA using the two-cycle amplification method of the Small Sample Target Labeling Protocol version II (Thermo Fisher Scientific), and total fragmented cRNA was hybridized to Human X3P GeneChip arrays (Thermo Fisher Scientific) for 16 h at 45 °C with constant agitation (60 rpm). Fluorescence was detected using an Affymetrix GeneChip Scanner 3000 7G (Thermo Fisher Scientific) and image analysis was performed with the Genechip Operating Software (Thermo Fisher Scientific) under the default settings. Microarray data were normalized by GCRMA of R (http://www.r-project.org/).
Transcriptome analysis
Gene set enrichment analysis (GSEA) [28, 29] was performed by the javaGSEA desktop application (ver. 4.0.1) using Hallmark gene sets (ver. 7.0). Gene sets with a false discovery rate (FDR) value < 0.05 after 1000 permutations were considered to be significantly enriched. Normalized enrichment scores (NESs) > 0 were considered upregulated, and subsystems with NESs < 0 were considered downregulated. Enrichment map was used for visualization of the GSEA results.
Differentially expressed genes (DEGs) between sCLs and glands structures were selected using an empirical criterion of a more than threefold change. Those genes expressed highly (averaged score > 1000) in the intracellular region were chosen. Strand NGS software (ver. 2.6; Agilent Technologies, CA, USA) was used to create heat maps.
3D culture model and co-culture with fibroblast
PLR123 or LS174T cells were suspended in 50% Matrigel (growth factor reduced, Corning, NY, USA) and dispensed into 24-well culture plates (250 cells/50 μL Matrigel droplet/well). After solidifying Matrigel at 37 °C, 650 μL of organoid culture medium (advanced DMEM/F12 medium supplemented with penicillin/streptomycin, 10 mM HEPES, 2 mM glutamax, 1× B-27 supplement, 1× N-2 supplement [Thermo Fisher Scientific], and 1 mM N-acetylcysteine [Merck, NJ, USA]) [30] was added and the cells were cultured at 37 °C under 5% CO2.
To observe the formation of sCL-like structures from cancer organoids, fibroblast co-cultures were analyzed. On day 10, the cancer organoids were collected through dissolution of Matrigel with Dispase I (FUJIFILM Wako Pure Chemical, Osaka, Japan). Then, the cancer organoids were transferred into 96-well plates precultured with CCD-18Co (1700 cells/well) for 3 days, or into Fibroblast Growth Medium 2 (PromoCell, Heidelberg, Germany) supplemented with 2.5% FBS on collagen I coated 96-well plates. The cultures were placed under a Confocal Quantitative Image Cytometer CQ1 (Yokogawa Electric, Tokyo, Japan) and bright-field images were monitored at 24 h intervals until day 4 (until day 11 for LS174T). The ratio of the formation of sCL-like structures was evaluated by analyzing 96 cancer organoids at day 4.
Whole mount IF staining
After removing the medium, PBS containing 4% PFA and 1% Triton X-100 was added and incubated for 2 h on ice, and further incubated in blocking buffer (BlockAid Blocking Solution, Thermo Fisher Scientific) for 2 h at room temperature. Primary antibodies (anti-LGR5 antibody [2L36, in-house] [11], Alexa Fluor 488-labeled anti-Ki67 antibody [BD Bioscience]) in blocking buffer were added and incubated overnight at 4 °C, then washed with 1% BSA/PBS and incubated for 1 h at room temperature with Alexa Fluor 555-labeled anti-mouse IgG2a antibody (Thermo Fisher Scientific). Finally, the wells were incubated with Alexa Fluor 650-phalloidin (Thermo Fisher Scientific) and DAPI for 40 min at room temperature. For optical clearing, the wells were incubated with SeeDB2G Solution 1 (1/3× Omnipaque350 [Daiichi-Sankyo, Tokyo, Japan] with 2% saponin [Nacalai Tesque, Kyoto, Japan]), Solution 2 (1/2× Omunipaque350 with 2% saponin), and Solution 3 (1× Omnipaque350 with 2% saponin) for 30 min each at room temperature, and with Solution 4 (Histodenz, Merch) overnight [31], and were observed with a confocal fluorescence microscope (A1, Nikon) with a ×10 objective lens (CFI Plan Apo λ, Nikon).
Results
Tumor hierarchy was reconstituted from LGR5-positive cells transplanted in NOG Mice
Xenograft tissues derived from LGR5-positive cells on day 3 to 19 after inoculation were evaluated under a microscope (Fig. 1a). As a whole, the structural shape changed from a small and simple cell cluster to a large and complicated structure over that period (Fig. 1b). In addition, we continued to observe the Matrigel until day 12. In this extended observation, we saw sCLs and glands known to be typical in moderate differentiated CRC. A few gland types were identified, including a structure with large lumen and the occasional stratified cell layer, and another structure consisting of many large glands or several glands connected back to back (Fig. 1c). sCLs with/without lumens emerged from day 3. On day 12, glands began to appear (Fig. 1d). The formation of tumor–mouse–stroma interface was observed on day 19.
a Schematic of the experiment. b Low magnification photographs of tumor tissue over time course. T tumor area. Scale bar = 100 μm. c Detailed structures in process of tumor formation on single section. Scale bars = 50 and 100 μm (rightmost image is gland/stroma interface). d Emergence of detailed structures over time. P presence. e Change in number of all tumor cells on one single plane of a section (n = 3, each point). f Change in number of LGR5-positive cells in 12 defined fields on one single plane of a section (n = 3, total). Blue: LGR5-positive; Red: LGR5-negative. g Immunofluorescence staining for LGR5 (green) and HLA-DMA (red). G gland; S small cluster. Rectangle shows small clusters. S1 shows LGR5-positive cells and S2 does not. Scale bar = 50 μm.
To analyze the relationship between tumor growth and LGR5-positive cells, we counted all of the tumor cells on maximum cut surface at day 3, 7, and 12 after inoculation. After day 7, the number of cells exponentially increased (Fig. 1e). At the same time, the number of LGR5-positive cells increased on a fixed inclination in the defined area (Fig. 1f), while LGR5-negative cells increased rapidly from day 7. It became clear that there were two phases in tumor growth from LGR5-positive cells; LGR5-positive cells mainly proliferate from day 3 to 7, and LGR5-negative cells from day 7 to 12. In other words, LGR5-negative cells overwhelmingly contribute to tumor growth compared with LGR5-positive cells. In addition, once we observed images stained by anti-LGR5 antibody, the difference in LGR5-positive cell distribution showed up even in morphologically identical sCLs (Fig. 1g). We suspected this was caused by the difference in 3D structure, so we attempted to reconstruct the 3D structures using serial section slides.
Serial sections revealed actual microscopic 3D structure of tumor
We tried to elucidate the microscopic 3D structure by observing the serial sections (Supplementary Fig. 1a–c). Structures were classified as either sCLs (with and without lumens) or glands; representative photos are shown in Supplementary Fig. 1c. Partial structures thought to be clusters in single sections were confirmed as sCLs in 3D. Although sCL-like structure appeared at the border of glands, its shape changed, becoming a clear ductal structure with a lumen in the middle of the tumor and resembling an sCL-like structure again in the end. This reconstructed 3D structure is shown at the bottom of Supplementary Fig. 1c. To accurately clarify these structures, the use of serial sections was necessary because the appearance changes with the depth of the section. By counting the sCLs without/with lumens and measuring their maximum diameters, we were able to continuously track them from day 3 to 12 (Supplementary Fig. 2). The average maximum diameter of sCLs without lumens was 22.8 μm (range 8–46 μm, n = 175), 18.1 μm (range 10–30 μm, n = 37), and 20.8 μm (range 6–40 μm, n = 26) at day 3, 7, and 12, respectively. sCLs with lumens at day 3 were relatively small (average 32.5 μm, range 18–60 μm, n = 52), but the average maximum diameter was 46.8 μm (range 20–74 μm, n = 42) and 47.2 μm (range 22–80 μm, n = 18) at day 7 and 12, respectively. sCLs without lumens were always smaller than those with lumens, indicating that the formation of a lumen is necessary to enlarge the structure.
sCLs contained LGR5-positive cells with high frequency
Next, we attempted to determine the exact distribution of LGR5-positive cells in each 3D structure. Pathological observation was performed on serial sections of double-stained samples for LGR5 and HLA-DMA, as are known markers of proliferating and nonproliferating CSC, respectively [11]. In sCLs without lumens, LGR5-positive cells were not dependent on the size of the structure but distributed throughout (Fig. 2a, b). In sCLs with lumens, ducts with LGR5-positive cells either distributed uniformly throughout the structure or scattered diffusely were observed (Fig. 2a, b). When both internal and external diameters were small, the frequency of LGR5-positive cells was high. Both were large, so their frequency was relatively low. In glands, there were ducts with both large and small lumen diameters, so the distribution of LGR5 was different depending on the location (Fig. 2a). Where the lumen was large, LGR5-positive cells were scattered diffusely, while a higher frequency of LGR5-positive cells were found in ducts with small lumens (Fig. 2a, arrow heads). Furthermore, an aggregation of LGR5-positive cells was found in part of the gland as protrusion of stratified cells, a so called “bud” (Fig. 2a, c, white arrow heads). In the sCLs, low expression of E-cadherin, distribution of β-catenin in nuclei, and low expression of Ki67 were observed (Fig. 2c). Furthermore, these three molecules showed the same expression pattern in the bud structure as well.
a Distribution of LGR5-positive cells in each 3D structure. Number shows the order of serial sections. Tumor cells are enclosed by dotted line. LGR5 (green) and HLA-DMA (red). White arrows indicate part of small lumen in gland. Scale bars = 25 μm for sCLs and 50 μm for glands. b Relationship between frequency of LGR5-positive cells and maximum external and internal diameter of sCLs. LGR5-positive grade: rare: white; occasional: blue; moderate: yellow; frequent: green. c Immunofluorescence for LGR5 (green)/E-cadherin (red), β-catenin (red), and LGR5 (green)/Ki67 (red) in sCLs and glands. Tumor cells are enclosed by dotted line. White arrows indicate bud structures in gland. Scale bar = 25 μm.
sCLs containing LGR5-positive cells continuously formed in the invasive front
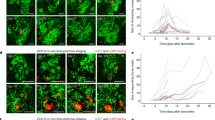
Histopathological observation during the late phase after inoculation was performed to confirm the continuous formation of sCLs, because it is possible that sCLs persist after inoculation during a limited observation period from day 3 to 12. At day 19, the interaction between the mouse interstitium and tumor was confirmed. In the center of the mass, there were differentiated structures composed of E-cadherin positive cells, and LGR5-positive cells were diffusely distributed (Fig. 3a). On the other hand, at the border of the mass, the invasive front, there were many LGR5-positive cells forming sCLs in a 3D manner (Fig. 3a, b). Moreover, long after inoculation, at day 35, the LGR5-positive signal was still more highly expressed in the invasive front than in the center (Fig. 3c, d). This result strongly suggests that sCLs highly expressing LGR5 form continuously in the invasive front and contribute greatly to the expansion of tumor size.
a Xenograft model inoculated with LGR5-positive CSC line (PLR123) at day 19 after inoculation. LGR5 (green) and E-cadherin (red). Multiple photos are combined into one image. Scale bar = 100 μm. b Serial sections for sCL in the invasive front at day 19. LGR5 (green) and HLA-DMA (red). Tumor cells are enclosed by dotted line. Number shows the order of serial sections. Scale bar = 50 μm. c Images for xenograft model inoculated with PLR123 line at day 35 after inoculation. LGR5 (green) and E-cadherin (red). Scale bar = 100 μm. d Histogram of LGR5 signal profile for individual samples at day 35 (left). Intensity of signal per 100 μm distance from tumor interface to center (0–500 μm) (right).
In addition, sCL-like structures were also observed in the primary site in a model where cells were engrafted into the cecum wall orthotopically (Supplementary Fig. 3a, b). Thus, the sCL is not only a structure that emerges in an artificial environment such as the subcutaneous region, but can also be observed in the intestine.
Formation of sCL from glands is reproduced in a 3D culture model using co-culturing system with fibroblasts as feeder cells
In order to confirm the formation of sCLs from glands in in vitro, a 3D culture model with fibroblasts as feeder cells was established (Fig. 4a). Because no sCL-like structures were observed after formation of ductal structures in the Matrigel culture [32], the need for some outside stimulation, such as from the stroma, was considered. After manipulating cancer organoids (day 10) from the Matrigel culture, these were transferred on to normal colon fibroblasts as feeder cells and kept for 4 days. Overall, 73 out of 96 cancer organoids with feeder cells had sCL-like structures surrounding the cancer organoid (Fig. 4b). In contrast, 24 out of 96 cancer organoids without feeder cells (only collagen coating) had sCL-like structures. In the cancer organoids with feeder cells, the edge of cancer organoid had a rough surface and some sCL-like cell aggregation began to bud as the cancer organoid became larger (Fig. 4c). Furthermore, the structure detached from main cancer organoid and grew into a new ductal structure (shown at high magnification at the bottom). The expression of LGR5 and Ki67 were confirmed in the sCL-like structures (Fig. 4d). In addition, the formation of sCL from glands was also confirmed in another cell line (LS174T) (Supplementary Fig. 4). Thus, the formation of sCLs from glands was confirmed in the 3D culture model, especially with fibroblast component.
a Schematic of the culture system. Scale bar = 50 μm. b Quantitative analysis for formation of sCL-like structures from main ductal structure. c Cancer organoid with feeder cells in time course. Rectangles indicate sCL-like structures detaching from main cancer organoid. Scale bar = 500 μm. d Expression of LGR5 (red), Ki67 (green), and phalloidin (white) in sCL-like structures. Scale bars = 200 μm (low) and 30 μm (high).
sCL has characteristics reflecting stress response and partial/hybrid EMT
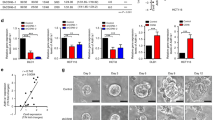
Next, we assessed the gene expression profile of sCL by microarray analysis for characterization. First, GSEA analysis was performed to evaluate which pathways were upregulated (Fig. 5a). In glands, pathways were upregulated such as E2F targets, oxidative phosphorylation, MYC targets, etc. It showed that cells could actively proliferate in glands. In contrast, in sCLs pathways were upregulated for stress response, such as with hypoxia, p53, and apoptosis pathways. In addition, sCLs were stimulated by TNFα and inflammatory responses in the environment. Furthermore, upregulation of EMT signature was noted, even though there was no typical upregulation of genes such as SNAIL, ZEB1, and TWIST [33].
a Gene set enrichment analysis (GSEA) using hallmark gene sets from the Molecular Signature Database. Statistically significant signatures were selected (FDR < 0.05) and placed in order of normalized enrichment score (NES). Red bars indicate the pathways enriched in the sCLs and blue bars those in the gland structures. GSEA enrichment plots are shown in the right. b Heatmap of differentially expressed genes (fold change > 3, averaged score > 1000). c Expression of LGR5 and S100A4 in xenograft sample of LGR5-positive CSC line at 12 days after inoculation. Expression of S100A4 was colocalized in a part of LGR5-positive sCLs (arrow heads). Scale bar = 100 μm.
To find genes highly expressed in sCLs, DEG analysis was conducted. Twenty-four genes were selected due to cytoplasmic localization, high expression, and threefold difference. The heatmap is shown in Fig. 5b. Upregulation correlated with prognosis for genes like S100A4 [34], AREG [35], CD55 [36], CEACAM6 [37], GPX2 [38] etc. in sCLs. S100A4 was the molecule with the highest difference in the expression list of Fig. 5b, and this was confirmed by IF for the xenograft sample at day 12 after inoculation (Fig. 5c). S100A4 was expressed in the same area of the sCLs as LGR5 (white arrow heads). These results suggest that sCL is biologically characterized by stress response and partial/hybrid EMT, and that it expresses prognosis-related genes.
sCLs with LGR5 expression exists in clinical CRC
To investigate the existence of sCLs with LGR5 expression in clinical samples, 29 cases of CRC were evaluated. The H-score for LGR5 is shown in Fig. 6a. Overall, 18 out of 29 cases expressed LGR5 (range; 0.2–44.0), and 8 out of the 10 cases with the highest expression had sCLs. In seven of these cases, LGR5 expression was higher in sCLs than in gland structures (Fig. 6b, Supplementary Fig. 5).
Discussion
A detailed process of growth from CSCs to tumors, including the actual distribution of LGR5, has yet to be fully elucidated, but LGR5 is still thought to be a driving force in a subset of human cancers [9]. According to the spatiotemporal observation of xenograft models in this study, tumor growth begins with the formation of an sCL consisting of LGR5-positive cells, then forms a lumen, which is still made up of LGR5-positive cells. Thereafter, the sCL grows larger, taking on LGR5-negative cells as proliferating tumor cells with polarity. During the process, a “bud” with a large accumulation of LGR5-positive cells forms as a structural component of glands, a new starting point for ductal formation. Then, it finally becomes multiple glands. In glands, this “bud” structure is similar to the sCL in the accumulation of LGR5-positive cells and the expression profiles of E-cadherinlow, β-cateninnuclei, and Ki67low.
It is possible for the “bud” in a gland to detach and become another starting point of the hierarchy. The sCLs formed continuously in the invasive front during tumor growth according to histopathological results for day 19 and 35 after inoculation. This strongly suggests that after detaching from glands, the formation of sCLs markedly contributes to tumor growth, as they are the starting point for glandular formation, are always found in the invasive front, and are at the frontline of tumor growth. Furthermore, our observation of the cancer organoids revealed that sCL-like structures detaching from glands were able to generate another ductal structure. These results confirm that sCLs are integral to the formation of the tumor hierarchy, as well as to the expansion of CSCs, as shown in Fig. 7. Indeed, some clinical CRC had sCLs with high expression of LGR5. It suggests that CSCs continuously expand through the formation of sCLs in some types of CRC, but not all. Further investigation is needed to determine which subpopulation of CRC patients exhibit this mode of CSC expansion.
In general, CSCs divide symmetrically to self-renew, or asymmetrically to give rise to CSCs and progenitors, which are committed to differentiate [3]. In this study, in the early phase of tumor growth from LGR5-positive cells and at the invasive front in the standard tumor growth process, we proved that the LGR5-positive cells population expands, accompanied by the formation of sCLs. For tumors, the advantages of an aggregation of cells over individual cells have been discussed [39]. Persistent epithelial cell–cell interactions within clusters can provide survival stimuli and potential resistance to anoikis [40], while the death of an isolated cell is thought to occur due to the loss of adhesion-dependent survival signals. This suggests that cells within sCLs are more resilient, able to avoid the pitfalls of isolated cells to more effectively expand CSCs.
Compared with glands, sCLs showed high expression of genes related to stress response and partial/hybrid EMT. sCLs consist of a small number of cells, and they enter (passively or actively) into a new microenvironment with interstitium. This microenvironmental change brings on physical stresses, oxidative stresses, and an immune assault that could induce apoptosis [39]. Thus, it is assumed that sCLs adapt to survive the severe microenvironmental change. The EMT signature was upregulated, but without the upregulation of typical EMT-related genes such as SNAIL, ZEB1, and TWIST [33] according to gene analysis. In addition, IF showed that sCLs had low expression of E-cadherin. Nowadays, it is thought that EMT do not represent all-or-none responses, but rather multistate processes, ranging from purely epithelial to purely mesenchymal via one or more intermediate phenotypes [41]. The intermediate phenotype of EMT is called the partial/hybrid EMT, and we think sCLs have the phenotype in this study. Further investigation will be needed, but this evidence implies sCLs may have a relationship to metastasis. In addition, the existence of sCLs with high expression of LGR5 was observed in primary and secondary sites in an orthotopic model using the same cell line (PLR123) [42]. This evidence also suggests that sCLs with high LGR5 expression contribute to metastasis.
TB and PDC are reported as prognosis factors in CRC [14,15,16,17]. Although the clinical significance of TB/PDC is recognized, the biology is still not fully understood [19, 43]. One of the major limitations is the absence of in vivo models [43]. TB is defined as a cell cluster consisting of less than five cells, and PDC as having five or more cells lacking evidence of glandular differentiation. Both kinds are mainly located in the invasive front on single sectioned slides [14,15,16,17]. It is thought that TB and PDC are sequential steps [44], but the distinction is arbitrarily defined [45]. In addition, when histopathologically evaluating TB in detail, it is necessary to confirm the 3D structure [22, 23], because the structure could be mischaracterized in 2D due to its small size. In this study, we defined the sCLs as a small aggregation of round or oval cells with or without a lumen, the diameter of which is less than ~100 μm in 3D. With a lumen, the structure is already beginning to differentiate, so it may be beyond the definition as a TB/PDC. However, we included sCLs with lumens in the same population those without, because they can both be recognized as sequential structures and because those with lumens still have a lot of LGR5-positive cells according to the histopathological evaluation. Thus, sCLs resemble TB/PDC morphologically, with its small size and lack of differentiation, even though it may not completely fit the definition of a TB\PDC.
Moreover, it is known that Ki67low, E-cadherinlow, and β-cateninnuclei are characteristic of TB [46], and this molecular similarity was also confirmed by IF. In addition, the similar biological characteristics of TB, such as its relationship to CSC [47] and partial EMT [16, 41], were revealed by the histopathological and gene analysis. Based on these results, the xenograft model inoculated tumor cells with CSC properties can recapitulate the formation of sCLs resembling TB/PDC. We believe that the xenograft model utilizing antibodies that detect CSCs can be an effective research tool for understanding TB/PDC.
Taken together, the process of tumor growth includes a CSC expansion cycle orchestrated by the continuous formation of sCLs, according to a xenograft model of moderate differentiated CRC. To obtain a deeper understanding of CSC function, we need to focus more on the process of tumor growth, including the behavior of CSCs. Understanding this process can lead to the discovery of new therapeutic targets against CSC.
References
Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91.
Cojoc M, Mäbert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Semin Cancer Biol. 2015;31:16–27.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34.
Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29.
Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427–32.
Leung C, Tan SH, Barker N. Recent advances in Lgr5 + stem cell research. Trends Cell Biol. 2018;28:380–91.
Gong X, Azhdarinia A, Ghosh SC, Xiong W, An Z, Liu Q. LGR5-targeted antibody–drug conjugate eradicates gastrointestinal tumors and prevents recurrence. Mol Cancer Ther. 2016;15:1580–90.
Junttila MR, Mao W, Wang X, Wang BE, Pham T, Flygare J, et al. Targeting LGR5 + cells with an antibody-drug conjugate for the treatment of colon cancer. Sci Transl Med. 2015;7:314ra186.
Morgan R, Mortensson E, Williams A. Targeting LGR5 in colorectal cancer: therapeutic gold or too plastic? Br J Cancer. 2018;118:1410–8.
Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H, Medema JP. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378–86.
Kobayashi S, Yamada-Okabe H, Suzuki M, Natori O, Kato A, Matsubara K, et al. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 2012;30:2631–44.
Yamazaki M, Kato A, Zaitsu Y, Watanabe T, Iimori M, Funahashi S, et al. Intensive immunofluorescence staining methods for low expression protein: Detection of intestinal stem cell marker LGR5. Acta Histochem Cytochem. 2015;48:159–64.
Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3:153–73.
Ueno H, Hashiguchi Y, Kajiwara Y, Shinto E, Shimazaki H, Kurihara H, et al. Proposed objective criteria for “Grade 3” in early invasive colorectal cancer. Am J Clin Pathol. 2010;134:312–22.
Lugli A, Kirsch R, Ajioka Y, Bosman F, Cathomas G, Dawson H, et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30:1299–311.
Zlobec I, Lugli A. Tumour budding in colorectal cancer: molecular rationale for clinical translation. Nat Rev Cancer. 2018;18:203–4.
Ueno H, Hase K, Hashiguchi Y, Shimazaki H, Yoshii S, Kudo SE, et al. Novel risk factors for lymph node metastasis in early invasive colorectal cancer: a multi-institution pathology review. J Gastroenterol. 2014;49:1314–23.
Karamitopoulou E. Tumor budding cells, cancer stem cells and epithelial-mesenchymal transition-type cells in pancreatic cancer. Front Oncol. 2013;2:1–5.
Mitrovic B, Schaeffer DF, Riddell RH, Kirsch R. Tumor budding in colorectal carcinoma: time to take notice. Mod Pathol. 2012;25:1315–25.
Gurzu S, Banias L, Kovacs Z, Jung I. Epithelial-mesenchymal transition of tumor budding in colorectal cancer: the mystery of CD44-positive stromal cells. Hum Pathol. 2018;71:168–9.
Yamada N, Sugai T, Eizuka M, Tsuchida K, Sugimoto R, Mue Y, et al. Tumor budding at the invasive front of colorectal cancer may not be associated with the epithelial-mesenchymal transition. Hum Pathol. 2017;60:151–9.
Bronsert P, Enderle-Ammour K, Bader M, Timme S, Kuehs M, Csanadi A, et al. Cancer cell invasion and EMT marker expression: a three-dimensional study of the human cancer-host interface: 3D cancer-host interface. J Pathol. 2014;234:410–22.
Prall F, Ostwald C, Linnebacher M. Tubular invasion and the morphogenesis of tumor budding in colorectal carcinoma. Hum Pathol. 2009;40:1510–2.
Sato Y, Mukai K, Watanabe S, Goto M, Shimosato Y. The AMeX method. A simplified technique of tissue processing and paraffin embedding with improved preservation of antigens for immunostaining. Am J Pathol. 1986;125:431–5.
Suzuki M, Katsuyama K, Adachi K, Ogawa Y, Yorozu K, Fujii E, et al. Combination of fixation using PLP fixative and embedding in paraffin by the AMeX method is useful for histochemical studies in assessment of immunotoxicity. J Toxicol Sci. 2002;27:165–72.
Marzouk O, Schofield J. Review of histopathological and molecular prognostic features in colorectal cancer. Cancers. 2011;3:2767–810.
Washington MK. Colorectal carcinoma: selected issues in pathologic examination and staging and determination of prognostic factors. Arch Pathol Lab Med. 2008;132:1600–7.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72.
Ke MT, Nakai Y, Fujimoto S, Takayama R, Yoshida S, Kitajima TS, et al. Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep. 2016;14:2718–32.
Fujii E, Yamazaki M, Kawai S, Ohtani Y, Watanabe T, Kato A, et al. A simple method for histopathological evaluation of organoids. J Toxicol Pathol. 2018;31:81–85.
Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription fators. Nat Cell Biol. 2019;21:102–12.
Kang YG, Jung CK, Lee A, Kang WK, Oh ST, Kang CS. Prognostic significance of S100A4 mRNA and protein expression in colorectal cancer. J Surg Oncol. 2012;105:119–24.
Ohchi T, Akagi Y, Kinugasa T, Kakuma T, Kawahara A, Sasatomi T, et al. Amphiregulin is a prognostic factor in colorectal cancer. Anticancer Res. 2012;32:2315–21.
Durrant LG, Chapman MA, Buckley DJ, Spendlove I, Robins RA, Armitage NC. Enhanced expression of the complement regulatory protein CD55 predicts a poor prognosis in colorectal cancer patients. Cancer Immunol Immunother. 2003;52:638–42.
Jantscheff P, Terracciano L, Lowy A, Glatz-Krieger K, Grunert F, Micheel B, et al. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic significance. J Clin Oncol. 2003;21:3638–46.
Liu D, Sun L, Tong J, Chen X, Li H, Zhang Q. Prognostic significance of glutathione peroxidase 2 in gastric carcinoma. Tumor Biol. 2017;39:1010428317701443.
Giuliano M, Shaikh A, Lo HC, Arpino G, De Placido S, Zhang XH, et al. Perspective on circulating tumor cell clusters: why it takes a village to metastasize. Cancer Res. 2018;78:845–52.
Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319–26.
Grigore A, Jolly M, Jia D, Farach-Carson M, Levine H. Tumor budding: the name is EMT. Partial EMT. J Clin Med. 2016;5:E51.
Kawai S, Yamazaki M, Shibuya K, Yamazaki M, Fujii E, Nakano K, et al. Three-dimensional culture models mimic colon cancer heterogeneity induced by different microenvironments. Sci Rep. 2020;10:3156.
Georges LMC, De Wever O, Galván JA, Dawson H, Lugli A, Demetter P, et al. Cell line derived xenograft mouse models are a suitable in vivo model for studying tumor budding in colorectal cancer. Front Med. 2019;6:139.
Reggiani Bonetti L, Barresi V, Bettelli S, Domati F, Palmiere C. Poorly differentiated clusters (PDC) in colorectal cancer: what is and ought to be known. Diagn Pathol. 2016;11:31.
Prall F. Tumour budding in colorectal carcinoma. Histopathology. 2007;50:151–62.
Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1:651–61.
Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–9.
Acknowledgements
We thank Yayoi Takai (Chugai Research Institute for Medical Science, Inc.) for help with pathological techniques, and Manami Kadowaki (Chugai Research Institute for Medical Science, Inc.) and Hiromichi Terashima (Chugai Pharmaceutical Co., Ltd) for microarray analysis. We are also grateful to Prof. Yoshihiko Maehara (Kyushu University) and Dr. Tatsumi Yamazaki (Chugai Pharmaceutical Co., Ltd) for their encouragement. Chugai Pharmaceutical Co., Ltd and Kyushu University have a collaborative research agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
MY, AK, EF, CK, NS, and MS are employees of Chugai Pharmaceutical Co., Ltd. KN, MN, TS, SK, EF, and MS are employees of Forerunner Pharma Research Co., Ltd. TW is an employee of Chugai Research Institute for Medical Science, Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yamazaki, M., Kato, A., Oki, E. et al. Continuous formation of small clusters with LGR5-positive cells contributes to tumor growth in a colorectal cancer xenograft model. Lab Invest 101, 12–25 (2021). https://doi.org/10.1038/s41374-020-0471-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-0471-y