Abstract
SRY (sex-determining region Y)-box 13 (Sox13), a member of group D of the SRY-related high mobility group (HMG) box (Sox) family, is a critical regulator of embryonic development and cartilage formation. Few studies have investigated the role of Sox13 in tumorigenesis. The present study reveals the clinical significance and biological function of Sox13 in hepatocellular carcinoma (HCC). First, the expression of Sox13 in HCC samples was evaluated by qRT-PCR and western blotting, and its association with clinicopathological features and prognosis was determined. We found that Sox13 expression was higher in tumor tissue than in paired nontumor tissue. The upregulation of Sox13 was associated with poor differentiation, metastasis, recurrence and poor overall, and tumor-free survival of HCC patients. The function of Sox13 on HCC cell migration and invasion was then assessed by Transwell assay, and the results demonstrated that Sox13 promoted HCC cell invasion, migration, and epithelial-to-mesenchymal transition (EMT). Notably, the invasion, migration, and EMT of HCC cells induced by Sox13 overexpression could be abolished by Twist1 depletion, and Sox13 was positively correlated with Twist1 at both the mRNA and protein levels. Mechanistically, we revealed that Sox13 activated Twist1 transcription and consequently upregulated Twist1 expression. Furthermore, Sox13 formed a heterodimer with Sox5, and this heterodimer functionally cooperated to enhance the transcriptional activity of Twist1. Our findings suggest that Sox13 serves as an oncogene in HCC, and might be a novel prognostic and therapeutic candidate.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is an aggressive type of cancer and a leading cause of cancer-associated mortality [1]. Due to the lack of obvious symptoms at its early stage, its high recurrence rate and resistance to chemotherapy and radiotherapy, HCC patients experience a very poor clinical outcome [1]. Thus, understanding the underlying signaling pathways and identifying key regulators in hepatocarcinogenesis could provide valuable insights into the molecular mechanisms of HCC and potential therapeutic targets for HCC.
The sex-determining region Y (SRY)-related high mobility group (HMG) box (Sox) genes are characterized by their HMG domain, which mediates DNA binding and bending [2]. The proteins encoded by Sox genes are pivotal transcription factors binding to the specific DNA sequence AACAAT and play key roles in cell fate decision and embryonic development [3]. In addition to functioning as transcription factors in the regulation of the development process, a growing number of studies have recently implicated Sox proteins in cancer progression [4, 5]. Different members of the Sox protein family play distinct roles in various types of cancers [6, 7]. More interestingly, a specific Sox protein can function as either a tumor suppressor or an oncogene in different human cancers. For instance, Sox9 is upregulated in HCC patients and considered a poor prognosis marker for these patients [8]. However, the overexpression of Sox9 inhibited melanoma growth [9]. The studies conducted to date have confirmed that at least ten members of the Sox family are involved in HCC development and progression. The overexpression of Sox4 [10], Sox5 [11], Sox9 [8], Sox12 [12], and Sox10 [13] has been detected in HCC and predicts a poor outcome for patients with HCC. In contrast to their oncogenic properties, decreased expression of Sox1 [13], Sox6 [14], Sox7 [15], Sox17 [16], and Sox11 [17] is relevant to the promotion of HCC progression, but the role of Sox13 in HCC has not yet been studied.
The Sox13 gene is localized in human chromosome 1q31.3–32.1. Sox13, together with Sox5 and Sox6, constitutes a class D subgroup of the Sox protein family and harbors two highly conserved functional domains [18]. One is the family-specific HMG box DNA-binding domain (aa 402–486), which is located in the C-terminal half of the protein, and the other is a group-specific coiled-coil domain (also called a leucine zipper motif, aa 126–203) located in the N-terminal half of the proteins [18]. The coiled-coil domain of SoxD proteins mediates both homodimerization and heterodimerization [18, 19]. Sox13 forms a homodimer through its leucine zipper motif, and this homodimerization reduces DNA binding [20]. Sox5-Sox6 heterodimerization enhances the binding of these proteins to DNA and is then activated by Sox9 at the Col2a1 enhancer [21].
This study provides the first insight into the function of Sox13 in HCC and demonstrates that increased Sox13 expression is correlated with metastasis and poor prognosis in HCC. We also show that Sox13 forms heterodimers and cooperates with Sox5 to enhance Twist1 transcriptional activity, and promotes HCC cell invasion and EMT. These results demonstrate that Sox13 is a novel oncogene in hepatocarcinogenesis and might be a potential therapeutic target for HCC.
Materials and methods
Human HCC samples, cell lines, and animals
Tumor samples were obtained from HCC patients at the time of surgery at Xiamen University Zhongshan Hospital. All the patients provided informed consent, and the study was approved by the Medical Ethics Committee of Xiamen University.
Human HCC cell lines (QGY-7701, PLC/PRF/5, Huh7, SMMC-7721, SK-Hep-1, HCC-LM3, and MHCC-97H), human 293T cell line and the immortalized normal liver cell line HL-7702 were used. MHCC-97H and HCC-LM3 cells were obtained from the Shanghai Cancer Institute. The other cell lines were purchased from the cell bank at the Shanghai Institute of Cell Biology (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, HyClone), 100 U/ml penicillin, and 100 U/ml streptomycin.
Antibodies
Anti-Sox13 (#18902–1-AP), anti-fibronectin (#15613-1-AP), anti-Sox5 (#13216-1-AP), and anti-actin (#66009-1-lg) antibodies were purchased from Proteintech. Anti-E-cadherin (#3195s), anti-vimentin (#5741s), and anti-N-cadherin (#13116s) antibodies were obtained from Cell Signaling Technology, and anti-Twist1 (#abs117807) antibody was purchased from Absin.
Dual-luciferase reporter assay
The Dual-Luciferase® Reporter (DLR™) Assay System (Promega, #E195A) was used to detect the luciferase activity. The relative luciferase activity was determined using Thermo Varioskan™ LUX, and the transfection efficiency was normalized by the Renilla luciferase activity.
Stable Sox13-knockdown and Sox13-overexpressing HCC cells
Two shRNA lentiviral plasmids and a lentiviral vector encoding human Sox13 and containing a puromycin resistance gene were constructed. The Turbofect transfection reagent (Thermo Scientific) was used for cell transfection. Recombinant lentivirus was produced by cotransfecting 293T cells with a mixture of the lentiviral and packaging plasmids, and the supernatant was then collected and used to infect the target cells.
Transwell assay
The transwell assay used to evaluate cell migration and invasion ability was performed as previously described [22].
Western blotting and qRT-PCR analysis
Western blotting and qRT-PCR were performed as previously described [22]. The primers used for qRT-PCR are shown in Table 1.
Immunohistochemistry assay
Paraffin-embedded tissues were serially sectioned at 5 µm and immunohistochemically examined using an antibody against Sox13. The Ultra-sensitive SP kit (Maxim) was then used for the detection of specific primary antibodies. The slides were visualized and photographed by light microscopy.
Co-IP assay
Cell lysates were extracted using lysis buffer with protease inhibitors, and immunoprecipitation was performed using a Dynabeads Immunoprecipitation Kit (Thermo Fisher). Briefly, the appropriate antibody and bead mixture was incubated in binding buffer at room temperature for 10 min and then incubated overnight with the cell lysate. The immunoprecipitate was then collected, washed, and examined by western blotting analysis. The immunoreactive products were visualized via enhanced chemiluminescence.
Statistical analyses
The data were presented as the means ± SD and analyzed using GraphPad Prism version 7. Student’s t test was used for the statistical analyses, and p < 0.05 was considered statistically significant.
Results
High Sox13 expression is associated with poor prognosis in HCC patients
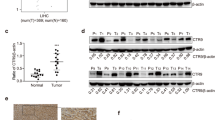
First, the mRNA level of Sox13 in paired HCC and normal tissues was detected, and the results showed that the Sox13 mRNA level was upregulated in tumors compared with the paired nontumor tissues (41/70 = 58.57%, p < 0.05, Fig. 1a). We then quantitatively analyzed the Sox13 mRNA expression level in HCC tissues from patients that experienced invasion and recurrence within 3 years postoperation. As shown in Fig. 1b, the patients who experienced invasion and recurrence had a markedly higher Sox13 mRNA level than those without invasion and recurrence respectively (p < 0.05, left and p < 0.01, right). The protein expression level of Sox13 in another set of 61 paired HCC tissues was then investigated by western blotting (Fig. 1c). A quantitative analysis showed that Sox13 protein expression was upregulated in 67.21% (41/61) of HCC samples (Fig. 1c). An immunohistochemical analysis further revealed that Sox13 protein expression was elevated in tumor tissues compared with adjacent nontumor tissues (Fig. 1d).
a Sox13 mRNA expression in 70 pairs of tumor and nontumor tissues. b Sox13 mRNA expression in tumor tissues from patients who did or did not experience invasion and recurrence. c Western blotting analysis of Sox13 protein expression in 61 pairs of tumor and nontumor tissues. N nontumor, T tumor. d Representative immunohistochemistry images of Sox13 expression in HCC and adjacent nontumor tissues. Scale bars = 100 μm (top) and 200 μm (below). e and f Kaplan–Meier analysis of overall survival (e) and tumor-free survival (f) in patients with upregulated (n = 41) and downregulated (n = 20) Sox13 expression. The difference in the overall and tumor-free survival rates between the two groups was significant.
Based on the Sox13 protein expression level detected by western blotting, we analyzed the correlation between Sox13 expression and clinicopathological features. The Sox13 protein level in older patients (24/29) was higher than that in younger patients (17/32, p = 0.014). More importantly, overexpression of Sox13 was significantly correlated with poor differentiation, metastasis, and recurrence (Table 2). Sox13 expression was not significantly associated with the patient gender, hepatitis B virus (HBV) level, tumor size, serum alpha-fetoprotein (AFP) level, or liver cirrhosis status (p > 0.05, Table 2). Subsequently, a Kaplan–Meier survival analysis was performed to assess the relationship between Sox13 protein expression and clinical outcomes in HCC patients (n = 61). Patients with high Sox13 expression exhibited significantly shorter overall survival times (Fig. 1e, p < 0.0001) and tumor-free survival times (Fig. 1f, p = 0.0003) than those with low Sox13 expression.
To further determine whether high Sox13 expression was related to the prognosis of HCC patients, the Cox proportional hazards regression model was performed. As shown in Table 3, the statistical data showed that high expression of Sox13 was an independent risk factor correlated with HCC patient prognosis [hazard ratio (HR) = 4.552 with 95% confidence interval (CI) = 1.231–16.837; p = 0.023]. These results demonstrated that Sox13 indicated the outcomes of HCC patients and might be a potential prognostic biomarker.
High Sox13 expression increases HCC cell migration and invasion
To investigate the oncogenic role of Sox13 in HCC, we first detected the expression of Sox13 in the normal liver cell line HL-7702 and seven HCC cell lines (QGY-7701, PLC/PRF/5, SMMC-7721, Huh7, SK-HEP-1, HCC-LM3, and MHCC-97H). As shown in Fig. 2a, b, both the mRNA and protein levels of Sox13 were higher in MHCC-97H and HCC-LM3 cells than in Huh7 and SMMC-7721 cells. We subsequently established stable Sox13-knockdown HCC-LM3 and MHCC-97H cells and Sox13-overexpressing SMMC-7721 and Huh7 cells. Western blotting assay was used to confirm the expression of Sox13 in these cell lines (Supplementary Fig. S1). The association between Sox13 expression and tumor metastasis indicated that Sox13 may be involved in HCC metastasis (Table 2). Then, the effect of Sox13 on the mobility of HCC cell lines was analyzed using the transwell assay. The results showed that the migratory and invasive capabilities of HCC cells were increased significantly by overexpression of Sox13 in SMCC-7721 and Huh7 cells (Fig. 2c, d), and clearly suppressed by knockdown of Sox13 in MHCC-97H and HCC-LM3 cells (Fig. 2e, f).
a and b mRNA (a) and protein (b) levels in normal liver cell and HCC cell lines. c and d Transwell assay analysis of migration and invasion of the control and Sox13-overexpressing SMCC-7721 (c) and Huh7 (d) cells. e and f Transwell assay analysis of migration and invasion of the control and Sox13-depleted MHCC-97H (e) and HCC-LM3 (f) cells. Scale bars = 100 μm.
Twist1 mediates Sox13-induced HCC cell EMT and mobility
To gain insights into the potential relationship of Sox13 with key transcription factors and signaling molecules implicated in epithelial-to-mesenchymal transition (EMT), EMT-related genes and transcriptional regulators were assessed by qRT-PCR. As shown in the Fig. 3a, knockdown of Sox13 upregulated the mRNA level of epithelial markers (E-cadherin) and downregulated the mRNA level of mesenchymal markers (N-cadherin, vimentin, and fibronectin) and EMT regulators (Twist1 and Snail). The opposite expression patterns for these markers were obtained in Sox13-overexpressing SMMC-7721 cells (Supplementary Fig. S2). The protein levels of E-cadherin, vimentin, N-cadherin, fibronectin, and Twist1 were regulated in a pattern consistent with observed changes in mRNA levels (Fig. 3b), suggesting that Sox13-induced EMT in HCC cells. We next investigated the underlying mechanism of Sox13 in regulating HCC metastasis and EMT. Twist1 was of particular interest to us because Twsit1 is the main gene that in response to Sox13 overexpression or depletion (Fig. 3a and Supplementary Fig. S2) and it has reported that Twsit1 can be transcriptionally activated by Sox5 [11, 23], another member of SoxD proteins. In addition, Twist1 is a key EMT inducer and critical for tumor metastasis, therefore, we then validated whether Twist1 is a key downstream target of Sox13 in promoting mobility and EMT in HCC cells. As shown in Fig. 3c, the overexpression of Sox13 increased HCC cell migration and invasion (p < 0.05), whereas the silencing of Twist1 in Huh7 Ctrl and Sox13-overexpressing cells dramatically abolished the Sox13-induced promotion of cell migration and invasion (p > 0.05). Meanwhile, we rescued the expression of Twist1 in LM3 shCtrl and Sox13-depleted cells. The restoration of Twist1 expression impaired the inhibition of cell migration and invasion induced by Sox13 depletion (Fig. 3d). Moreover, upregulation of Sox13 increased Twist1 expression and decreased E-cadherin expression, whereas knockdown of Twist1 expression significantly rescued the loss of E-cadherin expression induced by Sox13 overexpression (Fig. 3e). These results suggest that Twist1 is an essential regulator in the Sox13-mediated induction of HCC mobility and EMT.
a qRT-PCR analysis of the mRNA levels of EMT-related genes and regulators in Sox13-knockdown HCC-LM3 cells. b Western blotting analysis of Sox13, E-cadherin, vimentin, N-cadherin, fibronectin, and Twist1 protein expression in the indicated cells. c and d Twist1 was silenced in stable Sox13-overexpressing Huh7 cells (c) and overexpressed in stable Sox13-knockdown HCC-LM3 cells (d), then the migration and invasion of the indicated cells was assessed through a transwell assay (*p < 0.05). Scale bars = 100 μm. e Huh7 cells were stable overexpressed Sox13, and simultaneously infected with lentiviral particle expressing shRNAs targeting Twist1 to stably down-regulate Twist1 expression, and then the protein expression of Sox13, Twist1 and E-cadherin was detected in these cells by western blotting. Actin was loaded as an internal loading control.
Sox13 is positively correlated with Twist1 and activates Twist1 transcription
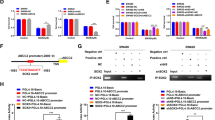
The above data show that Twist1 is involved in Sox13-mediated HCC metastasis. Then we analyzed the mRNA levels of Sox13 and Twist1 in 47 HCC tissues, and the results indicated a positive correlation between Sox13 and Twist1 expression in HCC samples (Fig. 4a, r = 0.493, p = 0.0004). This association was further confirmed by a western blotting analysis of 36 paired HCC tissues (Fig. 4b, c, r = 0.2927, p = 0.0126). Given that Sox13 is a transcription factor, we speculated that Twist1 might be a downstream target that is directly activated by Sox13. A dual-luciferase assay was then performed, and the data showed that the relative luciferase activity of Twist1 was increased in Sox13-overexpressing cells compared with the control cells (Fig. 4d). A sequencing analysis of the proximal promoter region of the Twist1 gene revealed three putative binding sites for Sox13 (Fig. 4e), and then we constructed fragments of the Twist1 promoter with different lengths. As shown in Fig. 4e, binding site 2 was critical for the Sox13-induced promotion of Twist1 transcriptional activity.
a Correlation coefficients were used to analyze the relationships between Sox13 and Twist1 mRNA expression levels in HCC tumor samples. b and c The expression of Sox13 and Twist1 proteins in tissues was detected by western blotting analysis. Representative data are shown (b), and their correlation was analyzed (c). N nontumor, T tumor. d Sox13 activated Twist1 transcriptional activity in HCC cells. The Twist1 luciferase reporter plasmid (−2825/+75) was transfected into Sox13-overexpressing Huh7 and SMMC-7721 cells, and the activity of the Twist1 promoter was detected using a dual-luciferase reporter assay. e Four luciferase reporter plasmids containing different binding sites were constructed, and a luciferase reporter assay was performed.
Sox13 and Sox5 cooperatively transactivate the activity of Twist1
Sox5 induces Twist1 transcriptional activity [23] and dimerizes with Sox6 through the coiled-coiled domain [18], and these findings prompted us to investigate whether Sox13 also dimerizes with Sox5. We detected Sox5 in the Sox13 immunoprecipitate but not the IgG immunoprecipitate from MHCC-97H cells (Fig. 5a), which suggested that Sox13 was able to dimerize with Sox5. We subsequently detected whether Sox5 and Sox13 can exhibit functional cooperation in activating the Twist1 promoter. First, we constructed a Sox13 mutant lacking a coiled-coiled domain (Sox13 ΔCC), which was unable to interact with Sox5. Then we performed a dual-luciferase reporter assay in 293T cells. As shown in Fig. 5b, the individual transfection of Sox13 or Sox5 modestly promoted the transcriptional activity of Twist1 (Lane 3 versus Lane 1 and Lane 4 versus Lane 1). The transcriptional activity of Twist1 in Sox13 ΔCC was decreased obviously compare with that in Sox13 wild type (Lane 3 versus Lane 2). Moreover, the cotransfection of Sox13 with Sox5 resulted in their cooperation to activate the Twist1 promoter (Lane 5 versus Lane 3 and 4). However, the transactive activity of Twist1 in Sox5 + Sox13ΔCC was much lower than that in Sox5 + Sox13 (Lane 6 versus Lane 5), which demonstrated that Sox13 ΔCC did not exhibit cooperativity with Sox5 in activating Twist1. Taken together, the Sox5–Sox13 interaction plays a role in the enhancement of Twist1 transcriptional activity.
Discussion
Sox family proteins fulfill their roles as transcription factors by controlling the expression of dozens of target genes at the transcription level by binding to discrete cis-regulatory elements [24]. These diverse combinations highlight the relevance of these transcription factors to many aspects of development, including sex determination, skeletogenesis, neurogenesis, and embryonic development [25]. In recent years, increasing evidence has revealed the involvement of different Sox proteins in tumor progression and metastasis [5, 25]. Unlike the other two SoxD members, Sox5 and Sox6, whose roles in malignant tumors have been widely studied [26], few studies have revealed the biological function of Sox13 in various tumors [27, 28]. The role of Sox13 in tumor progression, particularly in HCC, is currently not well understood.
Sox13 is highly expressed in oligodendroglioma [29] and U87 glioma-exposed endothelial cells (GECs), and has been found to be involved in the regulation of GEC angiogenesis [27]. Two recent studies demonstrated that Sox13 serves as an oncogene and regulates the proliferation of gastric cancer cells [28] and the metastasis of colorectal cancer (CRC) [30]. Our results indicated that high Sox13 expression in HCC was significantly associated with metastasis, recurrence, and poor prognosis. To our knowledge, this study provides the first disclosure of the relationship between Sox13 expression and the clinicopathological features of HCC patients.
The hallmarks of EMT include a decrease in epithelial markers (E-cadherin, Zo-1) and an increase in mesenchymal markers (vimentin, N-cadherin, and fibronectin) and EMT-related transcription factors (Twist1, Slug, Zeb1, and Snail), which restrain the function of E-cadherin [31]. Our data demonstrated that the knockdown of Sox13 upregulated E-cadherin expression but downregulated mesenchymal marker expression. Moreover, the migratory and invasive capabilities of HCC cells were weakened by Sox13 depletion and elevated by Sox13 overexpression, respectively. These findings provide a mechanistic framework for clarifying that HCC patients with high Sox13 expression exhibit higher rates of recurrence and invasion and a shorter survival time.
Twist1, a master regulator of EMT, is a well-known oncogene that mediates cell mobility [32]. Sox5 is elevated in several human malignancies, such as breast [23, 33], liver [11], lung [34] and gastric cancer [35], has been associated with worse patient survival, and promotes EMT and metastasis. The tumorigenicity of Sox5 is partially due to its capability to regulate Twist1 [11, 23, 35]. Sox12 transactivates fibroblast growth factor binding protein 1 (FGFBP1) and Twist1 expression to promote HCC migration and metastasis [12]. In this study, we also found that Sox13 clearly upregulated Twist1 expression, but the expression of other EMT transcription factors, such as Slug and Zeb1, was not significantly induced by Sox13. However, Sox13 can promote CRC metastasis by transactivating Slug and c-MET [30] and then upregulating the expression of Slug and c-MET. This difference in the results may in part be due to different in downstream effectors present in different cell types. Further depletion of Twist1 substantially impaired Sox13-induced HCC mobility and EMT, whereas the overexpression of Twist1 rescued the suppression of HCC cell mobility induced by Sox13 depletion, which demonstrated that Sox13 functions as an upstream modulator of Twist1 in HCC. Luciferase reporter assays demonstrated that Sox13 activated Twist1 activity at the transcriptional level and thereby upregulated Twist1 expression. Moreover, Sox13 expression was found to be significantly positively correlated with Twist1 expression at both the mRNA and protein levels.
Sox proteins usually exhibit gene regulatory functions by forming multiprotein complexes with other transcription factors [19, 36]. For example, Sox2 directly interacts with Oct3/4 via their DNA-binding domains and thereby forms a ternary complex to regulate FGF4 and UTF1 [37, 38]. The formation of Sox13-Hhex complex results in dislodging Sox13 from the Sox13-TCF1 complex and activates Wnt/TCF signaling in the early embryo [39]. In addition, SoxD proteins contain a coiled-coil domain that enables SoxD proteins to form homo and heterodimers [18], and it appears that the partner of a SoxD protein is a Sox protein belonging to the same group. A previous study demonstrated that the dimerization of Sox6 and Sox5 stabilizes their binding to adjacent DNA‐binding sites and that the resulting dimer cooperates with Sox9 to activate the Col2a gene [21]. In our study, we found that Sox13 interacted with Sox5 but not Sox12 (data not shown). Subsequently, a dual-luciferase reporter assay confirmed that the dimerization of Sox5 and Sox13 exerted a cooperative effect in activating Twist1 promoter activity (Fig. 5b). However, further studies are needed to elucidate the mechanisms through which Sox5 and Sox13 exert cooperative effects.
In summary, this study demonstrated that overexpression of Sox13 indicates the poor prognosis of HCC patients, and Sox13 serves as a key regulator of HCC cell migration, invasion, and EMT by transactivating Twist1 expression. Moreover, the Sox5–Sox13 interaction enhances the transcriptional activity of Twist1 (Fig. 6). Thus, these findings implicate Sox13 as a potential prognostic biomarker and therapeutic target for HCC.
References
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55.
Roose J, Korver W, Oving E, Wilson A, Wagenaar G, Markman M, et al. High expression of the HMG box factor sox-13 in arterial walls during embryonic development. Nucleic Acids Res. 1998;26:469–76.
Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–44.
Kumar P, Mistri TK. Transcription factors in SOX family: potent regulators for cancer initiation and development in the human body. Semin Cancer Biol. 2019;S1044-579X(18)30184-6.
Grimm D, Bauer J, Wise P, Kruger M, Simonsen U, Wehland M, et al. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2019;S1044-579X(18)30141-X.
Thu KL, Becker-Santos DD, Radulovich N, Pikor LA, Lam WL, Tsao MS. SOX15 and other SOX family members are important mediators of tumorigenesis in multiple cancer types. Oncoscience. 2014;1:326–35.
Zhu Y, Li Y, Jun Wei JW, Liu X. The role of Sox genes in lung morphogenesis and cancer. Int J Mol Sci. 2012;13:15767–83.
Guo X, Xiong L, Sun T, Peng R, Zou L, Zhu H, et al. Expression features of SOX9 associate with tumor progression and poor prognosis of hepatocellular carcinoma. Diagn Pathol. 2012;7:44.
Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, et al. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J Clin Investig. 2009;119:954–63.
Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31:1298–307.
Wang D, Han S, Wang X, Peng R, Li X. SOX5 promotes epithelial-mesenchymal transition and cell invasion via regulation of Twist1 in hepatocellular carcinoma. Med Oncol. 2015;32:461.
Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–33.
Zhou D, Bai F, Zhang X, Hu M, Zhao G, Zhao Z, et al. SOX10 is a novel oncogene in hepatocellular carcinoma through Wnt/beta-catenin/TCF4 cascade. Tumour Biol. 2014;35:9935–40.
Guo X, Yang M, Gu H, Zhao J, Zou L. Decreased expression of SOX6 confers a poor prognosis in hepatocellular carcinoma. Cancer Epidemiol. 2013;37:732–6.
Wang C, Guo Y, Wang J, Min Z. The suppressive role of SOX7 in hepatocarcinogenesis. PLoS ONE. 2014;9:e97433.
Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Epigenetics. 2010;5:743–9.
Liu Z, Zhong Y, Chen YJ, Chen H. SOX11 regulates apoptosis and cell cycle in hepatocellular carcinoma via Wnt/beta-catenin signaling pathway. Biotechnol Appl Biochem. 2019;66:240–6.
Lefebvre V. The SoxD transcription factors-Sox5, Sox6, and Sox13-are key cell fate modulators. Int J Biochem Cell Biol. 2010;42:429–32.
Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–7.
Kasimiotis H, Myers MA, Argentaro A, Mertin S, Fida S, Ferraro T, et al. Sex-determining region Y-related protein SOX13 is a diabetes autoantigen expressed in pancreatic islets. Diabetes. 2000;49:555–61.
Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–33.
Zhao B, Zhao W, Wang Y, Xu Y, Xu J, Tang K, et al. Connexin32 regulates hepatoma cell metastasis and proliferation via the p53 and Akt pathways. Oncotarget. 2015;6:10116–33.
Pei XH, Lv XQ, Li HX. Sox5 induces epithelial to mesenchymal transition by transactivation of Twist1. Biochem Biophys Res Commun. 2014;446:322–7.
Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6.
Xu YR, Yang WX. SOX-mediated molecular crosstalk during the progression of tumorigenesis. Semin Cell Dev Biol. 2017;63:23–34.
Liang Z, Xu J, Gu C. Novel role of the SRY-related high-mobility-group box D gene in cancer. Semin Cancer Biol. 2019;S1044-579X(19)30111-7.
He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J Exp Clin Cancer Res. 2019;38:65.
Bie LY, Li D, Wei Y, Li N, Chen XB, Luo SX. SOX13 dependent PAX8 expression promotes the proliferation of gastric carcinoma cells. Artif Cells Nanomed Biotechnol. 2019;47:3180–7.
Schlierf B, Friedrich RP, Roerig P, Felsberg J, Reifenberger G, Wegner M. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol. 2007;33:621–30.
Du F, Li X, Feng W, Qiao C, Chen J, Jiang M, et al. SOX13 promotes colorectal cancer metastasis by transactivating SNAI2 and c-MET. Oncogene. 2020;39:3522–40.
Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39.
Sun C, Ban Y, Wang K, Sun Y, Zhao Z. SOX5 promotes breast cancer proliferation and invasion by transactivation of EZH2. Oncol Lett. 2019;17:2754–62.
Chen X, Fu Y, Xu H, Teng P, Xie Q, Zhang Y, et al. SOX5 predicts poor prognosis in lung adenocarcinoma and promotes tumor metastasis through epithelial-mesenchymal transition. Oncotarget. 2018;9:10891–904.
You J, Zhao Q, Fan X, Wang J. SOX5 promotes cell invasion and metastasis via activation of Twist-mediated epithelial-mesenchymal transition in gastric cancer. Onco Targets Ther. 2019;12:2465–76.
Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–6.
Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–65.
Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–45.
Marfil V, Moya M, Pierreux CE, Castell JV, Lemaigre FP, Real FX, et al. Interaction between Hhex and SOX13 modulates Wnt/TCF activity. J Biol Chem. 2010;285:5726–37.
Acknowledgements
This work was supported by grants from the National Nature Science Foundation of China (81871963).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Feng, M., Fang, F., Fang, T. et al. Sox13 promotes hepatocellular carcinoma metastasis by transcriptionally activating Twist1. Lab Invest 100, 1400–1410 (2020). https://doi.org/10.1038/s41374-020-0445-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-0445-0