Abstract
The mechanisms which underlie defects in learning and memory are a major area of focus with the increasing incidence of Alzheimer’s disease in the aging population. The complex genetically-controlled, age-, and environmentally-dependent onset and progression of the cognitive deficits and neuronal pathology call for better understanding of the fundamental biology of the nervous system function. In this study, we focus on nuclear receptor binding factor-2 (NRBF2) which modulates the transcriptional activities of retinoic acid receptor α and retinoid X receptor α, and the autophagic activities of the BECN1–VPS34 complex. Since both transcriptional regulation and autophagic function are important in supporting neuronal function, we hypothesized that NRBF2 deficiency may lead to cognitive deficits. To test this, we developed a new mouse model with nervous system-specific knockout of Nrbf2. In a series of behavioral assessment, we demonstrate that NRBF2 knockout in the nervous system results in profound learning and memory deficits. Interestingly, we did not find deficits in autophagic flux in primary neurons and the autophagy deficits were minimal in the brain. In contrast, RNAseq analyses have identified altered expression of genes that have been shown to impact neuronal function. The observation that NRBF2 is involved in learning and memory suggests a new mechanism regulating cognition involving the role of this protein in regulating networks related to the function of retinoic acid receptors, protein folding, and quality control.
Similar content being viewed by others
Introduction
Memory loss marks the onset of mild cognitive impairment before the occurrence of dementia in Alzheimer’s disease. Understanding the mechanism of learning and memory loss is important both in the neurobiology of cognition and in the biology of neurodegenerative disease. It is then critical to identify potential novel regulators of learning memory and in this study, the focus was the nuclear receptor binding factor-2 (NRBF2) which we hypothesized could be an important modulator of protein quality control. In support of a role for NRBF2 in neuronal quality control it has been shown to bind to and modulate the autophagic activities of the BECN1–VPS34 complex [1,2,3,4]. Autophagy is a highly regulated cellular process, integrating signals ranging from nutrient availability to cellular stress and as we have recently proposed is a critical component of the antioxidant defenses of the cell [5]. Regulation of autophagy is especially important for post-mitotic neurons since these cells survive lengthy periods of time without cell division which prevents the dilution of accumulated toxic materials in proliferating cells. Furthermore, protein and organelle turnover is important for the quality control of intracellular functional components which can be damaged during normal electrophysiological and metabolic processes or environmental toxins and oxidative stress. It is then important to understand the mechanisms through which autophagy, as the key regulator of protein quality control, is controlled in neuronal cells. In addition, NRBF2 has been shown to modulate the transcriptional activities of retinoic acid receptor α (RARα) and retinoid X receptor α (RXRα) [6, 7]. It is then possible that NRBF2 could impact learning and memory through several mechanisms.
An important site for control of autophagic regulation is the autophagosomal membranes which contain phosphatidylinositol 3-phosphate (PI3P). This signaling molecule is produced by the kinase activity of class III PI3K, VPS34, through phosphorylating phosphatidylinositol. The association of BECN1 with VPS34 plays an important role in the nucleation of autophagosomes and can be regulated by transcription, post-translational modification, as well as interactions with regulatory proteins [8,9,10]. Beclin has been shown to interact with a number of proteins and plays a role in autophagy and cell survival [8,9,10,11]. Furthermore, both BECN1 and VPS34 knockout mice exhibit postnatal lethality or severe neuron loss in conditional knockouts, with mechanisms involving both autophagy deficits and endosomal trafficking [11,12,13]. We and others have previously demonstrated an essential role for VPS34 in PI3P production, endocytic and autophagic degradation, in diverse organs [12,13,14,15,16,17,18,19,20]. Recent studies using an autophagy interaction network based on proteomic analyses, confirmed by either Beclin or VPS34 pulldown assays, identified a novel BECN1/VPS34 regulator, NRBF2 [1,2,3,4]. The role the BECN1–VPS34 complex and potentially NRBF2 plays in regulating learning and memory is unclear but is plausible since BECN1 is decreased in Alzheimer’s disease brains and in particular microglia [21]. Furthermore, BECN1 heterozygous mice exhibited an exacerbated Alzheimer’s disease pathology in mouse models [22]. Further connecting the VPS34–BECN1 complex to cognition, it has been shown that knockout of the WDR45/WIPI4 protein, an effector of VPS34, in the nervous system, resulted in greatly impaired learning and memory [23].
In addition to interacting with the BECN–VPS34 complex, NRBF2 can also interact with RARα and RXRα [6, 7]. This is also an important alternative pathway regulating learning and memory, as evidenced by the observation that RAR/RXR deficiency causes learning and memory deficits in mouse models, and that retinoids or receptor agonists alleviate age-or disease-related memory deficits [24,25,26,27,28,29,30]. RAR/RXR receptors are present in mammalian nervous systems [31, 32], with levels decreasing with age [33, 34]. The blood levels of vitamin A and its derivatives in humans are also age and gender-dependent [35], and their deficiency in adult rodents results in learning and memory impairment [36,37,38].
These findings led us to the interesting hypothesis that NRBF2 plays a role in regulating learning and memory either through its effects of the autophagy process or the RAR/RXR system. NRBF2 whole body knockout mice can survive up to 12 months of age, while exhibiting smaller body weight at 4 months of age, and liver necrosis at 10 months [4]. These phenotypes potentially complicate a learning/memory study, especially at the age of 10 months or above. To overcome these challenges, we used a Nestin-cre:Nrbf2loxP/loxP strategy to engineer a knockout NRBF2 in the nervous system. These mice were viable with normal body weight and without obvious pathology. With young mice (6 months of age) we observed normal learning ability of the Nestin-cre:Nrbf2loxP/loxP mice in the Morris water maze hidden platform test, but impaired performance in probe trials. The learning deficits became strikingly profound in the hidden platform test at 12 months of age, consistent with an age-dependent effect, while appearance, body weight, and performances in open field and zero maze tests remain normal. The involvement of NRBF2 in learning and memory supports an important regulatory role for this protein with possible contributory mechanisms from autophagy and the RAR/RXR system.
Materials and methods
Generation of brain-specific NRBF2-deficient mice
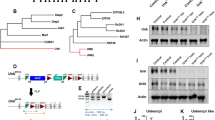
The mouse Nrbf2 gene is located on chromosome 10qB5.1 and contains four exons. The conditional deletion strategy for Nrbf2 gene was based on the deletion of 1924 bp sequence in exon 1. The Nrbf2 recombined locus was inserted with two loxP sites that flank exon 1 along with neomycin-resistant positive selection cassette flanked by flippase recognition target (FRT) sites. The diphtheria toxin A (DT-A) gene was used as a negative selection against random integration of the vector and inserted into downstream of the short arm. Homologous recombinant embryonic stem cells of C57BL/6 mouse origin were injected into C57BL/6J-Tyrc-2J/J blastocysts, and chimeric male mice were crossed with C57BL/6 Flp deleter female mice to excise the neomycin selection cassette. The resulting excised F1 mice were mated with C57BL/6 to produce Nrbf2f/+ mice. The progeny was again mated with Nestin-Cre transgenic mice expressing Cre recombinase that conditionally delete exon 1 of Nrbf2 gene in Nestin positive cells and produce Nrbf2f/f:Nestin-Cre mice (Fig. 1a). The following primers were used to detect wild-type Nrbf2 (a) and Nrbf2f (c) alleles:
a Genome structures of Nrbf2 endogenous locus and Nrbf2f/f:Nestin-Cre mice. (a) Nrbf2 endogenous locus. (b) Nrbf2 recombined locus with the insertion of neomycin selection cassette flanked by FRT and loxP sites. (c) Flp-mediated excision by breeding between recombined animals and ubiquitous Flp-recombinase expressing deleter mice enabled the deletion of FRT-flanked region. (d) Cre-mediated excision by breeding between recombined animals and Nestin-Cre transgenic mice, which enables the deletion of loxP-flanked region in Nestin-Cre expressing tissues. b Nrbf2 deficient mice (KO) exhibit diminished NRBF2 protein by western blot analyses compared to wildtype mice (WT) (postnatal day 25, n = 4–5 mice per genotype). Ctx: cortex, Cb: cerebellum, Str: striatum, MB: midbrain, HC: hippocampus. c NRBF2 protein levels appear normal in liver and heart in Nrbf2 deficient mice (KO) by western blot analyses compared to wildtype mice (WT) (postnatal day 25, n = 3 mice per genotype). Western blots of β-actin or GAPDH were used as loading controls.
nrbf1: 5′-CTCTCAATCCCTCCGCATCATCG-3′; nrbf2: 5′-GGTGCCTTTGCTTTAAGGCTCACG-3′.
cre1: 5′-TCGCGATTATCTTCTATATCTTCAG-3′; cre2: 5′-GCTCGACCAGTTTAGTTACCC-3′.
We have used n = 6–15 for behavioral studies from wildtype and NRBF2 deficient mice for each age group. All mouse experiments were done in compliance with the University of Alabama at Birmingham Institutional Animal Care and Use Committee guidelines.
Western blot analysis
Different regions of the mouse brain including cortex, cerebellum, striatum, midbrain, and hippocampus, as well as heart and liver, were collected for western blot analysis. Briefly, brains were dissected and homogenized in 1.5 ml centrifuge tubes in cell lysis buffer (50 mM Tris, 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid, 1% Triton X-100, pH 7.8) containing protease cocktail inhibitors (Roche). After 15 min on ice, samples were centrifuged at 1.3 × 104 rpm for 15 min. The supernatant was transferred into a fresh tube and pellet (Triton-X-100 insoluble fractions) were resuspended in 2% sodium dodecyl sulfate (SDS) in phosphate-buffered saline. Both supernatant and pellet fractions were used for bicinchoninic acid assay to determine protein quantification and 10–20 µg protein per lane were separated on a 12% SDS-polyacrylamide gel electrophoresis, then transferred to polyvinylidene difluoride membranes (Fisher Scientific, PI88520). Antibodies used were anti-NRBF2 (ProteinTech, 24858), anti-Synaptophysin (Cell Signaling, 5461), anti-PSD95 (Cell Signaling, 3450), anti-LC3 (Sigma, L8918), anti-p62 (Abnova, H00008878), anti-LAMP1 (Abcam, ab25245), anti-p-mTOR (Cell Signaling, 2971s), anti-mTOR (Santa Cruz, SC8319), anti-p-AKT (Cell Signaling, 9271), anti-AKT (Cell Signaling, 9272), anti-HSC70 (Abcam, Ab19136), anti-VPS34 (Sigma, V9764), and anti-BECN1 (Santa Cruz, sc11427). Anti-β-actin (Sigma, A5441) was used as a loading control. ECL reagent (Fisher Scientific, PIA34075), and ImageQuantTL (GE Healthcare) software were used to analyze western blot results.
Immunohistochemistry
Brains were placed in 10% buffered formalin (Fisher Scientific) overnight at 4 °C followed by paraffin embedding. Five-micrometer-thick sections were used for hematoxylin and eosin (H&E) and immunostaining. Both chromogenic and fluorescence immunohistochemistry were performed. Sections were treated with citrate buffer 0.01 M for 30 min at 90 °C. After antigen retrieval sections were blocked with 10% fetal bovine serum and 1% bovine serum albumin. The following primary antibodies were used: anti-GFAP (Invitrogen, 131-17719, 13-0300), anti-IBA1 (Abcam, ab5076), anti-NPAS4 (Invitrogen, MA5-27592), anti-STRA6 (Proteintech, 22001-1-AP), anti-STEAP2 (Invitrogen, PA5-20405), anti-HSPA5 (Invitrogen, PA5-34941), anti-SLC24A5 (Proteintech, 27747-1-AP), anti-EPS8L2 (Proteintech, 20461-1-AP), anti-LC3 (Sigma, L8918), anti-p62 (Abnova, H00008878), anti-VPS34 (Sigma, V9764), anti-Synaptophysin (Cell Signaling, 5461), and anti-BECN1 (Santa Cruz, SC11427), anti-PSD95 (Cell Signaling, 3450), and anti-Transferrin (Abcam, ab84036). The second antibodies for chromogenic DAB staining were: OneStep polymer HRP anti-rabbit (GeneTex GTX83399) or Immpress-HRP Horse Anti-Mouse TgG polymer Reagent (30027: Lot: ZF0604). The second antibodies for immunofluorescence staining were Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Cyanine3 (Invitrogen Cat # A10520) or Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (Invitrogen, A11004). DAB (3,3′-diaminobenzidine) kit (Vector SK-4100) was used in chromogenic stains. Nuclear counterstain was done by hematoxylin (Vector LS-J1042) and slides were mounted in Permount (Fisher, P36971). All immunofluorescence slides were nuclear counterstained and mounted using Invitrogen Prolong Diamond Antifade Mountant (Fisher, P36971). Keyence BZ-X microscope was used for capturing the immunostaining images.
TUNEL assay
Apoptotic cells were detected by TUNEL assay using an in situ cell death detection kit (Roche Diagnostic, 1176291910). Briefly, sections were subjected to antigen retrieval and then incubated with TUNEL assay mixture for 60 min at 37 °C in a humidified atmosphere in the dark. The method was tested in pathological sections where prior studies have detected TUNEL positive cells. TdT enzyme was omitted for negative controls.
Primary neuron cultures
Primary cortical neurons were cultured from P0 mouse pups. The mice were decapitated and brain was rapidly removed and dissections were performed immediately in ice-cold Hanks’ balanced sodium salts (without Ca2+ and Mg2+). After dissection cerebral cortices were chopped into small pieces and collected in a 15 ml Falcon tube containing HBSS. The tissues were further incubated for 30 min at 37 °C with papain (Worthington), then mechanically dissociated with a fire-polished Pasteur pipette. Cells were pelleted by centrifugation at 25 °C for 5 min at 1000 × g and resuspended in Neurobasal medium containing 2% B27 supplement (Invitrogen), 1% Pen-Strep (10,000 U/ml, 10,000 μg/ml) and 0.5 mM l-glutamine. Cells were then plated in 24-well or 6-well plates coated with 0.1 mg/ml poly-l-lysine (Sigma). The cultures were kept in a humid incubator (5% CO2, 37 °C).
Autophagic flux assay
For autophagic flux analysis, primary neurons were grown from Nrbf2f/fCre- and Nrbf2f/fCre+ pups, and at day 7 in vitro (DIV7) treated with and without 40 µM chloroquine (CQ) for 4 h, which blocks autophagy completion. Lysate was prepared from cells with and without CQ treatment, and western blot analysis for LC3 and p62 was performed.
Synaptosomal bioenergetics
Synaptosomes were prepared using the Percoll gradient method [39, 40]. The band between 10 and 23% Percoll represents synaptosomes. Fifteen micrograms of synaptosomes were aliquoted into glass-bottomed 96-well plates coated with polyethyleneimine and centrifuged at 3400g for 1 h at 4 °C, incubated with 100 nM MitoTracker for 30 min, and imaged. At this resolution, there was no difference in synaptosomal mitochondria amount between the two genotypes. Then 10 μg of synaptosomes were aliquoted into XF96 plates coated with polyethyleneimine and centrifuged at 3400g for 1 h at 4 °C. After resuspension with XF medium, the mitochondrial stress test was performed. Oxygen consumption rate (OCR) was measured at basal level and after sequential injections of Oligomycin (4 µg/ml), FCCP (2 µM), and antimycin A (10 µM). Basal (before oligomycin minus after antimycin), ATP-linked (before oligomycin minus after oligomycin), proton leak (after oligomycin minus after antimycin), maximal (after FCCP minus after antimycin), reserve capacity (maximal minus basal), and non-mitochondria (after antimycin) OCR were calculated as described previously [41,42,43,44,45,46,47].
In addition to mitochondrial stress test with endogenous nutrients, we also measured the function of individual mitochondrial complexes after permeabilization of the plasma membrane with PMP (Seahorse XF) bioscience proprietary reagents and providing specific substrates for each complex [48]. Oxidation of complex I linked substrates was measured by concurrent injection of PMP, pyruvate, malate, and uncoupler FCCP. This was followed by an injection of rotenone to inhibit complex I, and the addition of succinate (complex II substrate) to measure complex II substrate linked respiratory activities.
Complex IV substrate linked activities were determined in similar fashion by the addition of PMP, ascorbate, and tetramethyl-p-phenylenediamine (TMPD) with ADP. Rotenone was added to inhibit the possible reverse electron flow through complex I. Then the addition of azide was used to inhibit cytochrome c oxidase.
Behavioral tests
For the open field test [49, 50], data were recorded and analyzed using the Ethovision software. The mouse’s movements were tracked with regard to total distance traveled (cm), position inside the field (time in center versus against the wall in seconds), and mean velocity (cm/s).
For Zero Maze test, we used a circular maze (70 cm diameter) that was raised 40 cm above the table. The maze was divided into four equal parts, including two open areas and two closed areas. Mice were put at a starting point, and monitored for 4 min with an Ethovision camera-driven tracker system. The system recorded the position of the animal in the arena at 5 frames/s, and the time spent in each area was analyzed. Shorter time in the open area indicates increased anxiety.
A 3-day rotarod test [49] was performed in which the mice were placed on a rotating rod (San Diego Instruments) that gradually accelerated from 4 to 40 rpm over a 5 min period [51]. On each day mice were tested for three rounds at 1 h apart. The latency to fall was recorded in seconds for each trial and the average latency to fall was calculated for each mouse on each day. A decrease of latency suggests a decrease of motor coordination.
A water maze test was performed as described in detail before [49, 52, 53]. Briefly, we used a blue plastic pool with 120 cm in diameter, and a see-through round platform which is 10 cm in diameter and located 0.5 cm below the water surface. During day 1 through day 5 of the testing period, the mice were trained to find a hidden platform that was kept in a constant position throughout these 5 days. Four trials a day were run, and all starting positions in the four quadrants were equally used (in random order). The mice were given 60 s to find the platform and 10 s to stay on the platform. The inter-trial interval was approximately 2 min. Learning of the task was evaluated by recording the swimming speed, latency to find the platform, path length, and percentage of trials with which each animal found the platform. After the end of the four trials on day 5, the mice were tested in a 60 s probe trial (i.e., trial 21), i.e., with no escape platform present. Mice that had learned the platform position would predominantly search in the “correct” quadrant of the pool in the probe trial.
For a novel placement recognition test, mice were placed in a 40 × 40 × 60 cm3 (l × w × h) black Plexiglas box for 10 min to acclimate to the environment. The next day animals were placed in the same box with two identical objects for 3 min then returned to their cages. After a 2 h interval, the animals were returned to the box for a 3 min exposure to the same two objects from training but one object was placed in a new location. Exploratory behaviors consisted of the mouse’s nose coming in direct contact or within 1 cm of the object. Animals were excluded if they did not have at least 5 s of exploratory behavior or 1 s total exploration of each object.
For fear conditioning test, prior to the procedure, each animal was put in the conditioning chamber with a set of environmental cues for 2 min for habituation. Then three electrical foot shocks with paired cue were delivered. They remained in the chamber for an additional 30 s. Contextual memory was assessed 24 h later by re-exposing each animal to the same chamber for 5 min for evaluating freezing behavior. Cued test was assessed after a contextual memory test with a changed environment but with cues to determine amygdala dependent learning and memory. Percent freezing was calculated using Video Freeze Version 2.1.0 (Med Associates Inc.).
For examination of food intake, physical activity, and energy expenditure, over the course of the day, we used a comprehensive laboratory animal monitoring system (CLAMS). Mice were housed singly in CLAMS cages, for 1 week before measurements were made over 3 consecutive days in a 12:12 light:dark cycle; temperature was also maintained within the CLAMS. The system simultaneously measured food consumption, physical activity (movement through infra-red beams), and energy expenditure (through indirect calorimetry). The plastic flooring of the CLAMS cage has holes, allowing separation of feces and urine from the mouse (thereby ensuring accurate food intake assessments) [54,55,56].
Statistical analyses
All protein and mRNA results were normalized to internal control and then summarized as mean ± SEM; and the differences of means between wildtype and knockout mice were evaluated with Student t-test or Wilcoxon rank-sum test. All the outcomes from behavior studies were summarized as mean ± SEM. The t-tests were carried out to compare the genotype difference of the outcomes at each time point. For repeated measures, a linear mixed-effect regression model was conducted to evaluate the genotype difference over time. Results with a p-value of less than 0.05 were considered statistically significant. Experiments had a minimum of n = 3 mice for each group except a higher number of mice (n = 5–7 for open field, zero maze, rotarod, watermaze, and novel place recognition tests, n = 15 for fear condition) were used for behavioral studies.
RNAseq and data analyses
Hippocampi were dissected from five WT and five knockout (KO) male mice of 6 months of age. Preparation of total RNA and polyA sequencing library, RNAseq, and initial analyses were performed at the HudsonAlpha Institute for Biotechnology. Quality control of sequencing library included size verification and bioanalysis and quantitation by real-time PCR. NovaSeq S1 5-PE per lane yields 750 M reads, 75 Gb will yield ~50 M reads per sample. Transcriptome alignment used Edico’s Dragen System and the latest version of the mouse reference genome (mm10). For each sample, FeatureCounts was performed to get separate lists of counts for known genes and transcripts. To search for new transcripts, StringTie was performed and FPKM and TPM counts obtained. Further analysis was done using Linear Models for Microarray Analysis, and conserved transcription factor binding sites. Functional enrichment was done using the ToppFun application of the ToppGene Suite [57]. The p-value (FDR Benjamini–Hochberg corrected) was set at 0.05.
Quantitative real-time PCR analyses
RNA was prepared from the hippocampus using Trizol (Invitrogen). 0.5–2 µg of RNA was used to synthesize cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed with SYBR Green Mastermix (Thermo Fisher Scientific) with the following conditions: 95 °C, 5 min; (95 °C, 10 s; 60 °C, 10 s; 72 °C, 15 s) × 40 cycles. Results were normalized against an internal control (β-actin). Primers used are included in Supplementary Table 1.
Results
Generation of brain-specific conditional Nrbf2 deficient mice
We used the LoxP/Cre approach to generate conditional Nrbf2 deficient mice. We inserted LoxP sites by homologous recombination to flank exon 1 of mouse Nrbf2 gene in C57BL/6 embryonic stem cells (Fig. 1a) and injected into C57BL/6J-Tyrc-2J/J blastocysts. Breeding of chimeric mice with C57BL/6 Flp deleter mice resulted in an excised neomycin selection cassette. Germline transmitted Nrbf2f/+ mice were bred with Nestin-Cre transgenic mice to generate Nrbf2 nervous system-specific knockout (Nrbf2f/f:Nestin-Cre+) mice as evidenced by significantly decreased NRBF2 protein in wide areas of the brain (Fig. 1b). Western blot analyses of NRBF2 in the off-target tissues of liver and heart demonstrated that the levels of these proteins were not changed (Fig. 1c). The Nrbf2f/f:Nestin-Cre+ mice breed normally and have 5–8 pups per litter, continuously till 1.5 years of age, without apparent early mortality. The overall gross anatomy of the brain of the NRBF2 knockout mice was normal as assessed by H&E stained sagittal sections (Supplementary Fig. 1A–D).
We assessed LC3 and P62 levels in the hippocampus of 6 months old wildtype (WT) and NRBF2 knockout (KO) mice by western blot analyses. Results show that the levels of p62, BECN, and VPS34 were slightly elevated (Fig. 2), while the levels of LC3II, p-mTOR, total mTOR, p-AKT, total AKT, LAMP1, and HSP-70 were not significantly changed in KO mice (Supplementary Fig. 2A–E). There was a slight increase of LC3II/I (Supplementary Fig. 2A). Levels of the post-synaptic protein PSD95 was unchanged, while there was a modest increase in the pre-synaptic protein synaptophysin as determined by western blot analyses (Supplementary Fig. 3). Semi-quantitative immunohistochemistry analyses did not reveal remarkable changes in the levels of the post-synaptic protein PSD95, pre-synaptic protein synaptophysin, or LC3 in the hippocampus (Supplementary Fig. 4A–C). Autophagic flux was unchanged in primary cortical neurons in KO cells compared to control (Supplementary Fig. 5). Synaptosomal bioenergetics are normal at postnatal day 25 (Supplementary Fig. 6).
Mice were all at 6 months of age, protein extractions were from the dissected hippocampus (n = 3 mice per genotype). a Western blots and quantification of NRBF2 and p62. b Western blots and quantification of BECN and VPS34. All from n = 3 mice each genotype. Data = mean ± SEM normalized to control, Student t-test, *p < 0.05.
Neurological deficits in NRBF2 deficient mouse brain
To assess the neurological consequence of NRBF2 knockout (KO) in the nervous system, we performed behavior studies with open field and zero maze tests to assess spontaneous movement and anxiety (Supplementary Fig. 7A–D), mouse feeding, physical activity, and energy expenditures to assess whole body metabolism (Supplementary Fig. 7F), rotarod tests to assess motor coordination, Morris water maze, novel place recognition, and fear conditioning tests to assess learning and memory (Figs. 3 and 4). No significant alterations were observed in open field (Supplementary Fig. 7A, B) or zero maze (Supplementary Fig. 7C, D), both at 12 months and at 6 months of age between WT and knockout (KO) mice. Overall body weight (Supplementary Fig. 7E), mouse feeding, physical activity, and energy expenditures were unchanged at 6 months of age (Supplementary Fig. 7F).
All behaviors were performed with male mice. a, c 12 months of age (n = 5–7 each genotype). b, d 6 months of age (n = 5–7 each genotype). We found that the latency to fall, on the Rotarod test (49) with 4–40 rpm acceleration over 5 min, was not changed in 12 months old KO mice on all 3 days (p > 0.05) compared to WT (a), while decreased on the first day in 6 months old KO mice compared to WT mice, *p < 0.05 (b). c, d Total time on rod over the 3 testing days for 12 months old and 6 months old WT and KO mice also was of no statical difference.
All behaviors were performed with male mice. a–c, g 12 months of age (n = 5–7 each genotype). d–f 6 months of age (n = 5–7 each genotype). h, i, 2 months of age (n = 15 each genotype). a, d Morris Water Maze escape latency is plotted. KO mice exhibited longer escape latencies in multiple testing days both at 12 (a) and 6 months (d) of age compared to age-matched controls while the difference between the WT and the KO curve is bigger at 12 months of age. b, c, e, f KO mice exhibited less time spent in SE quadrant (b, e) and less SE location crossing (c, f) compared to WT mice in probe trial test, at both 12 (b, c) and 6 (e, f) months of age, while the differences between WT and KO mice are bigger at 12 months of age. The difference in time spent in the SE quadrant between WT and KO mice at 6 months of age (e) is not significant in Student t-test (p = 0.069). g KO mice are impaired in memory retention in the novel place recognition test at 12 months of age. During training both WT and KO mice explore objects in both locations equally. Testing was performed 2 h after training, during which KO mice exhibited no preference to the object placed at the novel place. h, i KO mice are impaired in memory retention in both the contextual (h) and the cued (i) fear conditioning test (2 months of age, n = 15 each genotype). Data = mean ± SEM, Student t-test, *p < 0.05 compared to control mice.
In the rotarod test, WT and knockout (KO) mice performed similarly at age of 12 months (Fig. 3a), while at age of 6 months, at day 1 there was a difference in performance between WT and KO mice (Fig. 3b). The mixed-effect regression model suggested that the overall time on rod combining all 3 days tended to be lower in KO mice at both 12 and 6 months of age, but did not reach statistical significance (p = 0.180 and p = 0.064, respectively) (Fig. 3c, d).
Significant deficits were observed in the Morris Water Maze at 12 months of age in knockout (KO) mice compared to WT mice as evidenced by escape latency in the hidden platform test (Fig. 4a) and less time spent in the SE quadrant (p = 0.0004 by two-tailed t-test and p = 0.009 by Wilcoxon rank-sum test) and fewer SE location crossing (p = 0.0002 by two-tailed t-test and p = 0.0081 by the Wilcoxon rank-sum test) during the probe trial (Fig. 4b, c).
Deficits were also observed in KO mice at 6 months of age, although the differences between WT and knockout (KO) mice were less pronounced (Fig. 4d–f). A linear mixed model suggested that on average the escape latency for KO mice was 12.4 s (95% CI 4.8–20.0, p = 0.0018) longer than WT mice; and the time decreased 5.0 s (95% CI 3.2–6.8, p < 0.0001) per day for both WT and KO mice. There was no significant difference in the trend between genotypes. The SE duration (p = 0.069 by two-tailed t-test, and p = 0.062 by Wilcoxon test) and the SE crossing frequency (p = 0.030 by two-tailed t-test, and p = 0.068 by Wilcoxon test) tended to be lower in KO mice.
To further characterize the learning and memory phenotype, we performed a novel place recognition test and found that the KO mice exhibit deficits in this test at 12 months of age compared to WT mice (Fig. 4g). In a larger cohort with n = 15 mice each, we also found that the KO mice exhibit deficits in fear conditioning at 2 months of age (Fig. 4h, i).
Significant decrease of expression of mRNA of SLC24A5 and EPS8L2, with significant accumulation of their respective proteins in the hippocampus of the NRBF2 knockout (KO) mice
Because of our observation that NRBF2 knockout (KO) primary neurons exhibit normal autophagic flux and NRBF2 knockout hippocampus only had subtle changes in autophagy protein levels, we further examined whether NRBF2 knockout has a significant impact in the hippocampal transcriptome. Hippocampi were dissected from 6 months old mice and subjected to RNAseq analyses. We confirmed that Nrbf2 gene expression was significantly decreased in the KO mice (to <1/36). From 24,306 genes analyzed, we found 47 whose expression levels had a >1.5-fold change (Supplementary Table 2). Pathway analyses using ToppFun application of the ToppGene Suite [57, 58] show that these genes are enriched for protein folding and quality control, as well as membrane vesicular proteins (downregulated), and immediate early genes/transcription factors (upregulated) (Fig. 5a). Real-time RT-PCR confirmed many of the genes identified as regulated by NRBF2, including decreases of EPS8l2 (epidermal growth factor receptor kinase substrate 8-like protein, implicated in cytoskeletal remodeling), SLC24a5 (solute carrier family 24, member 5, linked to ER and Golgi), HSPA5 (heat shock protein 5, linked to Alzheimer’s disease), PDIA4 (protein disulfide isomerase associated 4, linked to ER and Golgi), Steap2 (six-transmembrane protein of prostate 1, linked to ion transport and protein folding), and Stra6 (stimulated by retinoic acid 6); and an increase of TFRC (transferrin receptor) in NRBF2 knockout mice (Fig. 5b). LC3B, ATG14, and LAMP1 mRNAs did not change in NRBF2 knockout hippocampus. BECN mRNA is upregulated by ~20%. LAMP2a mRNA is trending down, whereas p62 mRNA is trending up.
a Upregulated genes (red/lighter circle nodes) enriched in transcription factors and downregulated genes (green/darker circle nodes) enriched in membrane proteins and protein folding are down. Enriched terms (p-value < 0.05 FDR B&H) are represented as blue rectangles. b Real-time RT-PCR (primers as listed in “Supplemental Table 1”) quantification of autophagy-lysosomal pathway genes and of NRBF2 target genes as revealed by RNAseq analyses (n = 3, 1 year of age). Upregulated in Nrbf2 KO include tfrc (transferrin receptor). Downregulated in Nrbf2 KO include eps8l2 (epidermal growth factor receptor kinase substrate 8-like protein, implicated in cytoskeletal remodeling), hspa5 (heat shock protein 5, linked to Alzheimer’s disease), pdia4 (protein disulfide isomerase associated 4, linked to ER and Golgi), slc24a5 (solute carrier family 24, member 5, linked to ER and Golgi), steap2 (linked to ion transport and protein folding), and stra6 (stimulated by retinoic acid 6).
Immunohistochemistry studies with TFRC, Steap2, HSPA5, and VPS34 did not show significant changes in the overall levels of these proteins (Supplementary Fig. 8). However, upon closer examination, we found that SLC24A5, STRA6, NPAS4d, and EPS8l2, as well as BECN1 and P62 proteins, exhibit an aggregate-like accumulation in the CA1 pyramidal/Radiatum/Lacunosum Moleculare layers, consistent with cell-specific disruption of protein quality control (Fig. 6).
Discussion
Since NRBF2 has been shown to interact with RXR/RAR transcription factors as well as the VPS34/BECN complex, we hypothesized that NRBF2 is important for learning/memory and tested this in a series of experiments using a novel mouse model with nervous system-specific NRBF2 knockout. We found that NRBF2 knockout mice exhibited normal physical activity, food intake, whole body energy expenditure, or metabolic reliance on the energy source (Supplementary Fig. 7). No change of synaptosomal bioenergetics was observed (Supplementary Fig. 6). By H&E staining, no evidence of neurodegeneration was observed at 6 months or 1 year of age (Supplementary Fig. 1). Strikingly, we observed significant learning and memory deficits in NRBF2 knockout mice, revealed by the Morris Water Maze, Novel Place Recognition, and fear conditioning test (Fig. 4). Interestingly we observed a decrease of freezing in both the contextual and cued fear conditioning test (Fig. 4). Importantly, normal behaviors in the open field and zero maze tests, were consistent with a lack of overt defects in movement or increased anxiety (Supplementary Fig. 7). However, there was a decrease in time spent on the rotarod in the first day of testing in 6-month-old knockout mice, and a trend for an overall decrease of motor coordination on the rotarod, consistent with potentially perturbed neuronal function (Fig. 3). Taken together these data as consistent with a specific deficit in both hippocampal mediated and amygdala mediated learning.
With regard to the potential involvement of altered autophagy and transcriptional function of NRBF2, we performed studies to compare autophagic homeostasis and transcriptomics in the hippocampus of the control and NRBF2 knockout mice. We found that in response to NRBF2 knockout, there was a modest elevation of P62, LC3II/I, BECN, and VPS34 proteins, with BECN mRNA also modestly elevated. LC3B and ATG14 mRNAs were unchanged. Thus the change of levels of BECN and possibly p62 may be due to both transcriptional activation and decreased turnover.
Despite the fact that prior studies indicating the role of NRBF2 in autophagic flux, we have not detected any changes in autophagic flux in primary neurons, with and without rapamycin. Neither have we detected any changes in p-mTOR or p-AKT levels in the brain. Autophagic flux in primary neurons is similar in WT and the knockout mice. This potentially can be due to the fact that the primary neurons are from postnatal day 0 mouse brains and that the western blot analyses are from adult brains. A proteotoxicity/quality control response to NRBF2 knockout will likely progress with age and not evident in the younger cells. Further, a change in levels of autophagy-related proteins does not always result in a change in autophagic flux. For example, other factors such as protein activities, whether they have become rate-limiting steps or not or whether other compensatory changes in the autophagy pathway have occurred may be important.
We have not detected any overt pathological changes in the knockout brain based on TUNEL staining and H&E staining. Furthermore, western blot analyses of PSD95 and synaptophysin proteins did not detect any changes in the knockout mice, suggesting a normal synaptic structure. This is not surprising as learning and memory can be altered with epigenetic, transcriptional, or other signaling changes in the brain [59,60,61]. Since NRBF2 has also been shown to interact with peroxisome proliferator-activated receptor alpha (PPAR-α), thyroid hormone receptor β (TRβ), retinoic acid receptor α (RARα), and retinoid X receptor α (RXRα) with the c-terminal 33 amino acids essential for the interaction in a yeast 2 hybrid system [6]. These interactions may influence NRBF2 function in the brain through an alternative mechanism to autophagy. Using RNAseq analyses, we found that the NRBF2 knockout altered the transcriptome in the mouse hippocampus. TFRC is upregulated by NRBF2 knockout, this may lead to increased cellular iron uptake and toxicity [62]. In human brains, TFRC is highly expressed in the hippocampus [63]. In mice, TFRC whole-body knockout is embryonic lethal due to severe anemia [58]. The aging model SAMP8 mice which also exhibit learning and memory deficits have a decreased Tfrc gene expression [64]. At present we do not know whether the alteration of its gene expression is a direct consequence of NRBF2 regulation of their transcription or indirect consequence downstream of NRBF2 deficiency. Additionally, it is unknown whether these changes contribute to the learning and memory deficits observed in NRBF2 knockout mice and whether they may be targeted to improve learning/memory.
Among the genes downregulated in the hippocampi of the NRBF2 knockout mice, Eps8l2 is involved in actin remodeling and its knockout leads to hearing loss [65, 66]. HSPA5 is a chaperone protein potentially implicated in Alzheimer’s disease [67]. HSPA5 transcription can be regulated by RXR::RAR and NRF2 [68, 69]. In addition to decreased TRFC and learning and memory deficits, the aging model SAMP8 mice also have increased HSPA8 and HSPA1A gene expression [64]. A decrease of Hspa5 mRNA has been shown in the hippocampus in aged cognitively impaired rats compared to young or aged cognitively unimpaired rats [70], and may result in misfolded proteins and decreased neuronal function. We do not know whether the change of its gene expression occurs in neurons versus populations of glia. Nonetheless, a recent single-cell transcriptomics study demonstrated that early changes in gene expression are cell type-specific while later changes in gene expression are shared among cell types and primarily involved in global stress response [71]. STRA6 is also a retinoic acid-responsive gene and may mediate cellular uptake of vitamin A and mediate RXR and RAR function [72]. Steap2 protein is a metalloreductase capable of spanning membranes, including the plasma membrane, vesicular tubular structures, and Golgi, and may be important for copper and iron homeostasis and protein trafficking [73, 74].
Considering that the NRBF2 knockout mice exhibit a profound deficit in learning and memory, it is important to understand how the downstream regulated genes are involved in the process. The upregulation of genes potentially can be compensatory to pathology or mediating pathology. In this regard, paradoxically, TFRC is upregulated in NRBF2 knockout brains and downregulated in AD brains [75], while Eps8l2 expression is downregulated in NRBF2 knockout brains while upregulated in APOE4/4 AD compared with APOE3/3 AD [76]. PDIA4 is a protein disulfide isomerase family protein important for protein quality control, and its expression is downregulated in NRBF2 knockout brains in this study while increased in AD brains [77]. Interestingly, and consistent with a protein quality control defect, we found aggregates of SLC24A5, EPS8L2, HPAS4, BECN1, and P62 in the CA1 Pyramidal/Radiatum/Lacunosum Moleculare layers. These may be due to a failure of quality control of these proteins, and may in turn contribute to altered synaptic physiology and cognitive deficits. SLC24A5 is a Na+/Ca2+–K+ exchanger previously shown to regulate pigmentation and localized to the trans-Golgi, while mitochondrial localization has also been noted recently [78,79,80]. EPS8L2 is an EPS8-like protein and is predicted to play a role in actin remodeling, extracellular vesicle production, and lysosome function [65, 66, 81, 82]. How these aggregates contribute to the learning and memory deficits will need to be further investigated. Recently, it has been reported that NRBF2 mRNA is downregulated in parahippocampal gyrus in human postmortem brains with Alzheimer’s disease compared to healthy controls, and that whole body NRBF2 knockout mice exhibit impaired memory, LTP, autophagy, and Aβ accumulation, further suggesting that a deficiency in NRBF2 may contribute to Aβ accumulation, dementia and cognitive deficits in Alzheimer’s diseases [83]. Our study provides insight into NRBF2 function in the nervous system and pathologies associated with its dysfunction. Taken together, we have shown that NRBF2 is important for learning and memory, and this may involve multiple interactive mechanisms. Future studies will determine whether these changes contribute to, or serve as an adaptive response that attenuates, the learning and memory deficits observed in NRBF2 knockout mice, and determine utility in dissecting neuronal versus glial function in cognition and dementia in the context of Alzheimer’s disease.
References
Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76.
Zhong Y, Morris DH, Jin L, Patel MS, Karunakaran SK, Fu YJ, et al. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem. 2014;289:26021–37.
Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J. 2014;461:315–22.
Lu J, He L, Behrends C, Araki M, Araki K, Jun WQ, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5:3920.
Giordano S, Darley-Usmar V, Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2014;2:82–90.
Yasumo H, Masuda N, Furusawa T, Tsukamoto T, Sadano H, Osumi T. Nuclear receptor binding factor-2 (NRBF-2), a possible gene activator protein interacting with nuclear hormone receptors. Biochim Biophys Acta. 2000;1490:189–97.
Flores AM, Li L, Aneskievich BJ. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J Investig Dermatol. 2004;123:1092–101.
Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists—mechanisms and experimental approaches. Redox Biol. 2015;4C:242–59.
Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80.
He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–9.
McKnight NC, Zhong Y, Wold MS, Gong S, Phillips GR, Dou Z, et al. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet. 2014;10:e1004626.
Wang L, Budolfson K, Wang F. Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience. 2011;172:427–42.
Zhou X, Wang L, Hasegawa H, Amin P, Han BX, Kaneko S, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci USA. 2010;107:9424–9.
Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagomez D, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol. 2013;190:5086–101.
Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci USA. 2012;109:2003–8.
Zhou X, Wang F. Effects of neuronal PIK3C3/Vps34 deletion on autophagy and beyond. Autophagy. 2010;6:798–9.
Bechtel W, Helmstadter M, Balica J, Hartleben B, Kiefer B, Hrnjic F, et al. Vps34 deficiency reveals the importance of endocytosis for podocyte homeostasis. J Am Soc Nephrol. 2013;24:727–43.
Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci USA. 2012;109:8670–5.
Morishita H, Eguchi S, Kimura H, Sasaki J, Sakamaki Y, Robinson ML, et al. Deletion of autophagy-related 5 (Atg5) and Pik3c3 genes in the lens causes cataract independent of programmed organelle degradation. J Biol Chem. 2013;288:11436–47.
McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. J Immunol. 2011;187:5051–61.
Lucin KM, O’Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, et al. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer’s disease. Neuron. 2013;79:873–86.
Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Investig. 2008;118:2190–9.
Zhao YG, Sun L, Miao G, Ji C, Zhao H, Sun H, et al. The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy. 2015;11:881–90.
Chiang MY, Misner D, Kempermann G, Schikorski T, Giguere V, Sucov HM, et al. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron. 1998;21:1353–61.
Etchamendy N, Enderlin V, Marighetto A, Vouimba RM, Pallet V, Jaffard R, et al. Alleviation of a selective age-related relational memory deficit in mice by pharmacologically induced normalization of brain retinoid signaling. J Neurosci. 2001;21:6423–9.
Ding Y, Qiao A, Wang Z, Goodwin JS, Lee ES, Block ML, et al. Retinoic acid attenuates beta-amyloid deposition and rescues memory deficits in an Alzheimer’s disease transgenic mouse model. J Neurosci. 2008;28:11622–34.
Nomoto M, Takeda Y, Uchida S, Mitsuda K, Enomoto H, Saito K, et al. Dysfunction of the RAR/RXR signaling pathway in the forebrain impairs hippocampal memory and synaptic plasticity. Mol Brain. 2012;5:8.
Kawahara K, Suenobu M, Ohtsuka H, Kuniyasu A, Sugimoto Y, Nakagomi M, et al. Cooperative therapeutic action of retinoic acid receptor and retinoid x receptor agonists in a mouse model of Alzheimer’s disease. J Alzheimers Dis. 2014;42:587–605.
Sodhi RK, Singh N. Retinoids as potential targets for Alzheimer’s disease. Pharmacol Biochem Behav. 2014;120:117–23.
Chakrabarti M, McDonald AJ, Will RJ, Moss MA, Das BC, Ray SK. Molecular signaling mechanisms of natural and synthetic retinoids for inhibition of pathogenesis in Alzheimer’s Disease. J Alzheimers Dis. 2016;50:335–52.
Krezel W, Kastner P, Chambon P. Differential expression of retinoid receptors in the adult mouse central nervous system. Neuroscience. 1999;89:1291–300.
Moreno S, Farioli-Vecchioli S, Ceru MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience. 2004;123:131–45.
Enderlin V, Alfos S, Pallet V, Garcin H, zais-Braesco V, Jaffard R, et al. Aging decreases the abundance of retinoic acid (RAR) and triiodothyronine (TR) nuclear receptor mRNA in rat brain: effect of the administration of retinoids. FEBS Lett. 1997;412:629–32.
Enderlin V, Pallet V, Alfos S, Dargelos E, Jaffard R, Garcin H, et al. Age-related decreases in mRNA for brain nuclear receptors and target genes are reversed by retinoic acid treatment. Neurosci Lett. 1997;229:125–9.
Breidenassel C, Valtuena J, Gonzalez-Gross M, Benser J, Spinneker A, Moreno LA, et al. Antioxidant vitamin status (A, E, C, and beta-carotene) in European adolescents—the HELENA Study. Int J Vitam Nutr Res. 2011;81:245–55.
Misner DL, Jacobs S, Shimizu Y, de Urquiza AM, Solomin L, Perlmann T, et al. Vitamin A deprivation results in reversible loss of hippocampal long-term synaptic plasticity. Proc Natl Acad Sci USA. 2001;98:11714–9.
Cocco S, Diaz G, Stancampiano R, Diana A, Carta M, Curreli R, et al. Vitamin A deficiency produces spatial learning and memory impairment in rats. Neuroscience. 2002;115:475–82.
Etchamendy N, Enderlin V, Marighetto A, Pallet V, Higueret P, Jaffard R. Vitamin A deficiency and relational memory deficit in adult mice: relationships with changes in brain retinoid signalling. Behav Brain Res. 2003;145:37–49.
Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J Neurochem. 2009;109:1179–91.
Flynn JM, Choi SW, Day NU, Gerencser AA, Hubbard A, Melov S. Impaired spare respiratory capacity in cortical synaptosomes from Sod2 null mice. Free Radic Biol Med. 2011;50:866–73.
Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, et al. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 2011;51:1621–35.
Giordano S, Dodson M, Ravi S, Redmann M, Ouyang X, Darley-Usmar VM, et al. Bioenergetic adaptation in response to autophagy regulators during rotenone exposure. J Neurochem. 2014;131:625–33.
Schneider L, Giordano S, Zelickson BR, Johnson S, Benavides A, Ouyang X, et al. Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic Biol Med. 2011;51:2007–17.
Benavides GA, Liang Q, Dodson M, Darley-Usmar V, Zhang J. Inhibition of autophagy and glycolysis by nitric oxide during hypoxia-reoxygenation impairs cellular bioenergetics and promotes cell death in primary neurons. Free Radic Biol Med. 2013;65:1215–28.
Higdon AN, Benavides GA, Chacko BK, Ouyang X, Johnson MS, Landar A, et al. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am J Physiol Heart Circ Physiol. 2012;302:H1394–409.
Mitchell T, Johnson MS, Ouyang X, Chacko BK, Mitra K, Lei X, et al. Dysfunctional mitochondrial bioenergetics and oxidative stress in Akita+/Ins2-derived beta-cells. Am J Physiol Endocrinol Metab. 2013;305:E585–99.
Giordano S, Lee J, Darley-Usmar VM, Zhang J. Distinct effects of rotenone, 1-methyl-4-phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS ONE. 2012;7:e44610.
Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat Protoc. 2014;9:421–38.
Crabtree D, Boyer-Guittaut M, Ouyang X, Fineberg N, Zhang J. Dopamine and its metabolites in cathepsin D heterozygous mice before and after MPTP administration. Neurosci Lett. 2013;538:3–8.
Xu M, Guo Y, Vorhees CV, Zhang J. Behavioral responses to cocaine and amphetamine administration in mice lacking the dopamine D1 receptor. Brain Res. 2000;852:198–207.
Meyer OA, Tilson HA, Byrd WC, Riley MT. A method for the routine assessment of fore- and hindlimb grip strength of rats and mice. Neurobehav Toxicol. 1979;1:233–6.
Slane JM, Lee HS, Vorhees CV, Zhang J, Xu M. DNA fragmentation factor 45 deficient mice exhibit enhanced spatial learning and memory compared to wild-type control mice. Brain Res. 2000;867:70–9.
Zhang J, McQuade JM, Vorhees CV, Xu M. Hippocampal expression of c-fos is not essential for spatial learning. Synapse. 2002;46:91–9.
Brewer RA, Collins HE, Berry RD, Brahma MK, Tirado BA, Peliciari-Garcia RA, et al. Temporal partitioning of adaptive responses of the murine heart to fasting. Life Sci. 2018;197:30–9.
Bray MS, Ratcliffe WF, Grenett MH, Brewer RA, Gamble KL, Young ME. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes. 2013;37:843–52.
Tsai JY, Villegas-Montoya C, Boland BB, Blasier Z, Egbejimi O, Gonzalez R, et al. Influence of dark phase restricted high fat feeding on myocardial adaptation in mice. J Mol Cell Cardiol. 2013;55:147–55.
Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11.
Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet. 1999;21:396–9.
Zeng H, Chattarji S, Barbarosie M, Rondi-Reig L, Philpot BD, Miyakawa T, et al. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–29.
Sarinana J, Tonegawa S. Differentiation of forebrain and hippocampal dopamine 1-class receptors, D1R and D5R, in spatial learning and memory. Hippocampus. 2016;26:76–86.
Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J, et al. Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep. 2016;16:2666–85.
Altamura S, Muckenthaler MU. Iron toxicity in diseases of aging: Alzheimer’s disease, Parkinson’s disease and atherosclerosis. J Alzheimers Dis. 2009;16:879–95.
Hanninen MM, Haapasalo J, Haapasalo H, Fleming RE, Britton RS, Bacon BR, et al. Expression of iron-related genes in human brain and brain tumors. BMC Neurosci. 2009;10:36.
Armbrecht HJ, Siddiqui AM, Green M, Farr SA, Kumar VB, Banks WA, et al. SAMP8 mice have altered hippocampal gene expression in long term potentiation, phosphatidylinositol signaling, and endocytosis pathways. Neurobiol Aging. 2014;35:159–68.
Dahmani M, Ammar-Khodja F, Bonnet C, Lefevre GM, Hardelin JP, Ibrahim H, et al. EPS8L2 is a new causal gene for childhood onset autosomal recessive progressive hearing loss. Orphanet J Rare Dis. 2015;10:96.
Furness DN, Johnson SL, Manor U, Ruttiger L, Tocchetti A, Offenhauser N, et al. Progressive hearing loss and gradual deterioration of sensory hair bundles in the ears of mice lacking the actin-binding protein Eps8L2. Proc Natl Acad Sci USA. 2013;110:13898–903.
Arispe N, De Maio A. Memory loss and the onset of Alzheimer’s disease could be under the control of extracellular heat shock proteins. J Alzheimers Dis. 2018;63:927–34.
Shi W, Xu G, Wang C, Sperber SM, Chen Y, Zhou Q, et al. Heat shock 70-kDa protein 5 (Hspa5) is essential for pronephros formation by mediating retinoic acid signaling. J Biol Chem. 2015;290:577–89.
Ahn B, Pharaoh G, Premkumar P, Huseman K, Ranjit R, Kinter M, et al. Nrf2 deficiency exacerbates age-related contractile dysfunction and loss of skeletal muscle mass. Redox Biol. 2018;17:47–58.
Haberman RP, Colantuoni C, Stocker AM, Schmidt AC, Pedersen JT, Gallagher M. Prominent hippocampal CA3 gene expression profile in neurocognitive aging. Neurobiol Aging. 2011;32:1678–92.
Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7.
Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–5.
Routhe LJ, Moos T. Handling iron in restorative neuroscience. Neural Regen Res. 2015;10:1558–9.
Ji C, Kosman DJ. Molecular mechanisms of non-transferrin-bound and transferring-bound iron uptake in primary hippocampal neurons. J Neurochem. 2015;133:668–83.
Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS ONE. 2011;6:e16266.
Xu PT, Li YJ, Qin XJ, Scherzer CR, Xu H, Schmechel DE, et al. Differences in apolipoprotein E3/3 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Neurobiol Dis. 2006;21:256–75.
Montibeller L, de Belleroche J. Amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD) are characterised by differential activation of ER stress pathways: focus on UPR target genes. Cell Stress Chaperones. 2018;23:897–912.
Zhang Z, Gong J, Sviderskaya EV, Wei A, Li W. Mitochondrial NCKX5 regulates melanosomal biogenesis and pigment production. J Cell Sci. 2019;132:jcs232009
Ginger RS, Askew SE, Ogborne RM, Wilson S, Ferdinando D, Dadd T, et al. SLC24A5 encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem. 2008;283:5486–95.
Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–6.
Offenhauser N, Borgonovo A, Disanza A, Romano P, Ponzanelli I, Iannolo G, et al. The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol Biol Cell. 2004;15:91–8.
Welsch T, Younsi A, Disanza A, Rodriguez JA, Cuervo AM, Scita G, et al. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res. 2010;316:1914–24.
Lachance V, Wang Q, Sweet E, Choi I, Cai CZ, Zhuang XX, et al. Autophagy protein NRBF2 has reduced expression in Alzheimer’s brains and modulates memory and amyloid-beta homeostasis in mice. Mol Neurodegener. 2019;14:43.
Acknowledgements
The authors thank members of Dr. Zhang’s laboratory for technical assistance and discussions, HudsonAlpha and Dr. Dan Dorset for RNAseq analyses, Drs. Thomas Van Groen, Jeremy Day, Jeremy Herskowitz, and Andrew Arrant for their help with learning and memory behavioral and related studies, and Drs. Terry L. Lewis and Marissa Menard (Molecular Detection Core and Cellular and Molecular Neuropathology Core) for assisting with immunohistochemistry. The authors thank UAB bridge funding, UAB CIRC for Next-Generation Sequencing Pilot Research Grant Program for supporting the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ouyang, X., Ahmad, I., Johnson, M.S. et al. Nuclear receptor binding factor 2 (NRBF2) is required for learning and memory. Lab Invest 100, 1238–1251 (2020). https://doi.org/10.1038/s41374-020-0433-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-0433-4