Abstract
SOX2 is recognized as an oncogene in human small cell lung cancer (SCLC), which is an aggressive neuroendocrine (NE) tumor. However, the role of SOX2 in SCLC is not completely understood, and strategies to selectively target SOX2 in SCLC cells remain elusive. Here, we show, using next-generation sequencing, that SOX2 expressed in the ASCL1-high SCLC (SCLC-A) subtype cell line is dependent on ASCL1, which is a lineage-specific transcriptional factor, and is involved in NE differentiation and tumorigenesis. ASCL1 recruits SOX2, which promotes INSM1 and WNT11 expression. Immunohistochemical studies revealed that SCLC tissue samples expressed SOX2, ASCL1, and INSM1 in 18 out of the 30 cases (60%). Contrary to the ASCL1–SOX2 signaling axis controlling SCLC biology in the SCLC-A subtype, SOX2 targets distinct genes such as those related to the Hippo pathway in the ASCL1-negative, YAP1-high SCLC (SCLC-Y) subtype. Although SOX2 knockdown experiments suppressed NE differentiation and cell proliferation in the SCLC-A subtype, they did not sufficiently impair the growth of the SCLC-Y subtype cell lines in vitro and ex vivo. The present results support the importance of the ASCL1–SOX2 axis as a main subtype of SCLC, and suggest the therapeutic potential of targeting the ASCL1–SOX2 axis.
Similar content being viewed by others
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide [1]. Small cell lung cancer (SCLC) accounts for ~14% of all lung cancers and is genetically considered to be one of the most aggressive malignant neuroendocrine (NE) tumors [2]. The development of novel target molecules in therapies for SCLC remains limited. The findings of basic studies on the molecular mechanisms underlying small cell carcinogenesis have yet to be clarified, and advances in novel therapeutic development are expected [3].
Recently, Rudin et al. proposed a nomenclature to describe SCLC subtypes according to the dominant expression of transcription factors (TFs); they divided SCLC into four subtypes, which were considered to be the master regulators of SCLC: achaete–scute complex homolog 1 (ASCL1) (SCLC-A), neurogenic differentiation factor 1 (NEUROD1), yes-associated protein 1 (YAP1) (SCLC-Y), and POU class 2 homeobox 3 (POU2F3) [4]. SCLC-A subtype belongs to NE high SCLC, which could be associated with the increased expression of ASCL1, a member of the basic helix–loop–helix family of TFs [5]. In contrast, SCLC-Y subtype belongs to NE low SCLC, which is associated with the activation of NOTCH, Hippo, and RE1 silencing TF (REST) genes [6, 7]. ASCL1 was previously shown to be expressed at a high frequency in SCLC [8] and plays a pivotal role in SCLC tumorigenesis as an oncogene [9]. In human SCLC, SOX2 was also recognized as an oncogene [10], along with ASCL1.
The major aim of the present study is to elucidate the mechanisms by which SOX2 affects the phenotype and heterogeneity of SCLC. We hypothesized that SOX2 may contribute to distinct transcriptional programs and biological characteristics in both the SCLC-A and SCLC-Y subtype. To test this hypothesis, the present study was designed to investigate the following: (1) a comparison of the target genes of SOX2 in human SCLC cell lines by chromatin immunoprecipitation (ChIP) sequence (ChIP-seq) analysis; (2) the relationship between ASCL1 and SOX2 in SCLC cell lines, surgically resected tissues, and public datasets of SCLC; and (3) the functional difference in SOX2 between the SCLC-A and SCLC-Y subtype cell lines. We herein demonstrated that SOX2 regulates distinct transcriptional programs in both the SCLC-A and SCLC-Y subtype, and that in the SCLC-A subtype, SOX2 functions in an ASCL1-dependent manner.
Materials and methods
Cell lines
Seven SCLC cell lines (H69, H889, SBC1, H69AR, H1688, SBC3, and SBC5), three adenocarcinoma (ADC) cell lines (A549, H358, and H1975), and two squamous cell carcinoma (SCC) cell lines (H2170 and H226) were used in the present study. H69, H889, H69AR, H1688, A549, H358, H1975, H2170, and H226 were purchased from ATCC (Manassas, VA, USA), and SBC1, SBC3, and SBC5 from the Japan Collection of Research Bioresources Cell Bank (Osaka, Japan). Cell lines that were used in the experimental studies for ChIP, RNA sequence (RNA-seq), and transfection were authenticated by comparison with the database of JCRB Cell Bank.
Tissue samples
We obtained tissue samples of SCLCs (n = 30), ADCs (n = 20), and SCCs (n = 20) from the lung cancer files of the Department of Pathology and Experimental Medicine and resected at the Department of Thoracic Surgery of Kumamoto University from 70 patients for this study. Histological diagnoses of the samples were made according to the criteria of the WHO [11]. These sections were used for immunohistochemical (IHC) staining. The present study followed the guidelines of the Ethics Committee of Kumamoto University.
Western blotting (WB) analysis
Cells were prepared for WB analysis as previously described [12]. A list of the primary antibodies used is shown in Table 1.
Chromatin immunoprecipitation
Three SCLC cell lines (H69, H889, and SBC3), the A549 ADC cell line, and the ASCL1-transfected A549 cell line were used in the present study. Cells were fixed in 1% formaldehyde (Thermo-Fisher) in PBS at room temperature for 10 min. Crosslinked cells were lysed with LB1 (50 mM HEPES-KOH (pH7.5), 140 mM NaCl, 1 mM EDTA, 10% (w/v) glycerol, 0.5% (w/v) NP-40, 0.25% (w/v) TritonX-100, proteinase inhibitor cocktail) (Sigma) and washed with LB2 (10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, proteinase inhibitor cocktail). Chromatin lysates were prepared in RIPA buffer (Thermo-Fisher; 25 mM Tris-HCl pH7.6, 150 mM NaCl, 0.1% SDS, 1% NP-40, 1% sodium deoxycholate, proteinase inhibitor cocktail), sonicated with Covaris S220 (peak incident power, 175; acoustic duty factor, 10%; cycle per burst, 200; treatment time, 600 s; cycle, 6). ChIP was performed using chromatin lysates equivalent to 1.0 × 107 cells, and protein A Dynabeads (Thermo-Fisher) coupled with the antibody against Sox2 (developed in our group). The specificity of the anti-Sox2 antisera used in this study was previously verified in a previous paper using Sox2-null ES cells [13], and the cross-reactivity to other Sox family members was also previously verified [14]. After 4 h of incubation at 4 °C, beads were washed four times in a low salt buffer (20 mM Tris-HCl (pH 8.0), 0.1% SDS, 1% (w/v) TritonX-100, 2 mM EDTA, 150 mM NaCl) and two times in a high salt buffer (20 mM Tris-HCl (pH 8.0), 0.1% SDS, 1% (w/v) TritonX-100, 2 mM EDTA, 500 mM NaCl). Chromatin complexes were eluted from the beads by agitation in elution buffer (10 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5 mM EDTA, 1% SDS) and incubated overnight at 65 °C for reverse crosslinking. Eluates were treated with RNase A and proteinase K, and DNA was ethanol precipitated.
ChIP-seq data analysis
ChIP-seq libraries were prepared using 20 ng of input DNA, and 1–3 ng of ChIP DNA with KAPA Library Preparation Kit (KAPA Biosystems) and NimbleGen SeqCap Adapter Kit A or B (Roche) and sequenced by Illumina NextSeq 500 (Illumina) using Nextseq 500/550 High Output v2 Kit (Illumina) to obtain single end 75-nt reads. The resulting reads were trimmed to remove the adapter sequence and low-quality ends using Trim Galore! v0.4.3 (cutadapt v.1.15). The trimmed ChIP-seq reads were mapped to the UCSC hg38 genome assemblies using Bowtie2 v2.3.3 with default parameters. The resulting SAM files were converted to BAM format using SAMtools v1.5. Peak calling was performed using MACS2 v2.1.1. with input DNA as a control including a q-value cut-off of 0.01 for SOX2 ChIP-seq. The distance to the nearest transcription start site (TSS) and gene feature of the peaks were obtained from Ensembl human annotation data (GRCh38) using the annotatePeakInBatch of ChIPpeakAnno and biomaRt R packages. Peaks in the gene body were first annotated with the option ‘output=”overlapping”, and the remaining peaks were then annotated to the nearest TSSs regardless of the distance between them. Protein binding sites were shown along with genomic loci from RefSeq genes on the genome browser integrated genome viewer (IGV). Motif identification was performed using MEME-ChIP v5.1.1 website (http://meme-suite.org/tools/meme-chip) [15]. The sequence of ±50 bp around the peak summits were obtained from UCSC hg38 whole genome annotation data using the ChIPpeakAnno R package, and masked for repetitive elements using RepeatMasker version open-4.0.7. For motif database, we selected “eukaryote DNA vertebrates (in vivo and in silico)”.
RNA sequence
RNA-seq was performed by the Liaison Laboratory Research Promotion Center (LILA) (Kumamoto University) as follows. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Germany) and 2100 Bioanalyzer was used to detect the concentration and purity of total RNA. All samples with an RNA integrity number > 7.5 were used for sequencing. DNA libraries prepared according to the Illumina Truseq protocol using Truseq Standard mRNA LT Sample Prep Kits and sequenced by Nextseq 500 (Illumina) were used for analysis, and the data were converted to Fastq files. Quality control of the data was performed by FastQC. The reads were then trimmed to remove the adapter sequence using Trim Galore! v0.5.0 (Cutadapt v 1.16), and low-quality reads were filtered out using FASTX-toolkit v0.0.14. The remaining reads were aligned to the Ensembl GRCh38.93 reference genome using STAR ver.2.6.0a. FPKM (fragments per kilobase of exon per million reads mapped) values were calculated using Cuffdiff. Then, differential expression analysis was performed using the ExAtlas website (https://lgsun.irp.nia.nih.gov/exatlas/).
Gene ontology (GO) analysis
The Database for Annotation, Visualization, and Integrated Discovery Bioinformatics Resources 6.7 (DAVID 6.7, http://www.david.niaid.nih.gov) was used for the GO analysis [16]. We used the default analysis settings on the website. The gene lists contained significant genes in the RNA-seq analysis and were also targeted by SOX2 in our ChIP-seq analysis (Supplementary Table S1). SOX2-targeted plus the highly expressed 346 genes in the SCLC-A subtype had been combined for DAVID analysis in SCLC-A subtype. Similarly, SOX2-targeted plus the highly expressed 825 genes in the SCLC-Y subtype had been combined for DAVID analysis of SCLC-Y subtype (Fig. 1b, c). The uncategorized genes for GO analysis were eliminated in each subtype. The top ten categories of biological processes in each SCLC-A and SCLC-Y subtype were taken as excerpts for Fig. 1c.
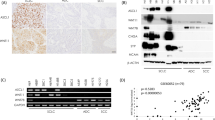
a Western blotting (WB) analysis of SOX2, ASCL1, INSM1, and YAP1 in lung cancer cell lines, including small cell lung carcinoma (SCLC), adenocarcinoma (ADC), and squamous cell carcinoma (SCC). SOX2 was more strongly expressed in all SCLC cell lines than in the NSCLCs examined. Four out of the seven SCLC cell lines simultaneously expressed ASCL1 and INSM1. β-ACTIN served as an internal control. b ChIP-seq and RNA-seq were conducted to analyze putative SOX2 target genes and mRNA expression in the H69, H889, and SBC3 cell lines. As shown in the Venn plot, 346 specific SOX2-bound genes, which also significantly expressed mRNAs in the SCLC-A subtype, were identified (shared in NCI-H69 and NCI-H889). The top 20 highest expressed genes from our RNA-seq dataset are presented. INSM1 was included as a common gene. In a similar manner, 825 specific SOX2-bound genes that also significantly expressed mRNAs were identified in the SCLC-Y subtype of the SBC3 cell line. Hippo pathway-related genes and REST were included as common genes (Supplementary Table S1). c Results of the gene ontology (GO) functional analysis. The top ten enriched categories in biological processes in each of the SCLC-A and SCLC-Y subtype are shown. We used the DAVID online bioinformatics tool for GO functional analysis and extracted the top ten enriched categories in biological processes from each subtype. Neuron-related categories were more likely to be included in the SCLC-A subtype compared to cell development- or movement process-related categories in the SCLC-Y subtype (Fig. 1c). The distinct functions of SOX2 in each subtype are suggested.
Transfection with RNA interference (RNAi)
We purchased Sox2 RNAi (sc-41120) and Ascl1 RNAi (sc-37692) from Santa Cruz Biotechnology (Santa Cruz, USA) and transfected them into cells using an electroporator (NEPA21 pulse generator; Nepa Gene, Chiba, Japan) as described in the manufacturer’s instructions. These were a pool of 3 different RNAi duplexes and sequences for Sox2 were as follows. sense; 5′-GAAUGGACCUUGUAUAGAUTT-3′, antisense; 5′-AUCUAUACAAGGUCCAUUCTT-3′ (sc-38408A), sense; 5′-GGACAGUUGCAAACGUGAATT-3′, antisense; 5′-UUCACGUUUGCAACUGUCCTT-3′ (sc-38408B), and sense; 5′-GAAUCAGUCUGCCGAGAAUTT-3′, antisense; 5′-AUUCUCGGCAGACUGAUUCTT-3′ (sc-41120C). The sequences for Ascl1 were as follows. sense; 5′-CCAACAAGAAGAUGAGUAATT-3′, antisense; 5′-UUACUCAUCUUCUUGUUGGTT-3′ (sc-37692A), sense; 5′-GAAGCGCUCAGAACAGUAUTT-3′, antisense; 5′-AUACUGUUCUGAGCGCUUCTT-3′ (sc-37692B), sense; 5′-GUUCGGGGAUAUAUUAAGATT-3′, antisense; 5′-UCUUAAUAUAUCCCCGAACTT-3′ (sc-37692B). Control siRNA (Cat# sc-36869) was used as a control. Cells were harvested 48–72 h post transfection.
Cell counting assay
Cells were prepared for cell counting assays as previously described [17]. H69, H889, SBC1, H69AR, and SBC3 cells were used in this study.
IHC and evaluation
Formalin-fixed, paraffin-embedded specimens, which were cut into 4-μm-thick sections and mounted onto MAS-GP-coated slides (Matsunami Glass Ind., Osaka, Japan), were used and are detailed in our previous study [17]. A list of the primary antibodies used is shown in Table 1.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using an RNeasy Mini Kit (Qiagen) in accordance with the manufacturer’s instructions and detailed in our previous study [17]. A list of the primers used is shown in Table 2.
SOX2-knockout experiment using SBC3 and H69AR cell lines
Genome editing using CRISPR/Cas9 was used for knockout of the SOX2 gene in the SBC3 and H69AR cell lines. We purchased SOX2-knockout vectors from GeneCopoeiaTM (Rockville, USA). These were three different human SOX2 sgRNA/Cas9 all-in-one expression clones (NM_003106.2). The sgRNA target sequences of SOX2 were as follows: ATGGGCCGCTTGACGCGGTC (HCP217628-CG10-3-10-a), CGCCCGCATGTACAACATGA (HCP217628-CG10-3-10-b), and ATTATAAATACCGGCCCCGG (HCP217628-CG10-3-10-c). CCPCTR01-CG10 (GeneCopoeiaTM) was used as a control. The sgRNA target sequence was GGCTTCGCGCCGTAGTCTTA. Regarding the establishment of SOX2-knockout H69AR cells, we obtained pSpCas9(BB)-2A-Puro(px459) from Addgene (Cambridge, MA) [18]. The sgRNA target sequences of SOX2 were ATAATAACAATCATCGGCGG, GACCGCGTCAAGCGGCCCAT, and ACAGCCCGGACCGCGTCAAG.
Tumor xenotransplantation experiment
Eight-week-old male Rag2(−/−):Jak3(−/−) mice (a generous gift from Prof. S. Okada; Kumamoto University) were used. Two groups of mice were subcutaneously injected; one group was injected with 2 × 106 stably transfected cells with ASCL1, and the other group was injected with an equal number of the control cell population. After 4 weeks, the animals were anaesthetized with an overdose of intraperitoneal pentobarbital (30 mg/kg body weight), and sacrificed by cutting the inferior vena cava. The subcutaneous tumors were removed and fixed. The samples were fixed with 10% formalin and embedded in paraffin. Tissue sections were stained with hematoxylin and eosin and additional sections were used for IHC staining. To investigate the function of SOX2 in the SBC3 cell line (a SCLC-Y subtype), a total of 1.0 × 106 cells from the mock-transfected or SOX2-knockout SBC3 cell lines were subcutaneously injected into the backs of mice (nine animals each). Twenty days after the first injection, tumors were removed and measured. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines (#A2019-034).
Statistical analysis
Spearman’s correlation coefficient (ρ) was calculated for the correlation analysis. Cell counting data were expressed as the means ± standard deviation of triplicate measurements. Differences in mean values between the groups were analyzed by a two-tailed statistical analysis using the Student’s t test. GraphPad Prism version 7.04 (San Diego, CA, USA) was used for all statistical analyses. p values less than 0.05 were considered to be significant.
Results
SOX2 is overexpressed and targeted distinct genes between SCLC-A and SCLC-Y subtype cell lines
To examine SOX2 expression patterns, we performed WB analysis of 12 lung cancer cell lines (7 SCLCs, 3 ADCs, and 2 SCCs). WB analysis revealed that the SOX2 protein was generally expressed at markedly higher levels in all examined SCLC cell lines than in the NSCLC cell lines. Four out of the seven SCLC cell lines simultaneously expressed SOX2, ASCL1, and insulinoma-associated protein 1 (INSM1) (Fig. 1a). INSM1 is a crucial regulator of NE differentiation in lung cancer, and is specifically expressed in SCLC, along with ASCL1 [19]. In the present study, H69, H889, SBC1, and H1688 belonged to the SCLC-A subtype cell lines, which were positive for ASCL1 and INSM1. In contrast, H69AR, SBC3, and SBC5 were classified as the SCLC-Y subtype cell lines, which were negative for ASCL1 but highly positive for YAP1 (Fig. 1a). We then conducted ChIP-seq to analyze SOX2 target genes in the H69, H889, and SBC3 cell lines, followed by motif analysis (Supplementary Figs. S1 and S2). We also combined the results of the RNA-seq analysis for these cell lines, which showed the global expression levels of mRNAs. As shown in a Venn plot, we identified 346 SOX2-bound genes (shared in NCI-H69 and NCI-H889) that were specifically occupied in the SCLC-A subtype and expressed at higher levels in the SCLC-A subtype than in the SCLC-Y subtype (Fig. 1b). Some neuron-related genes, such as INSM1, seizure related 6 homolog like, or synaptic vesicle glycoprotein 2B (SV2B), were included as their common target genes. On the other hand, we identified 825 SOX2-bound genes (targeted in SBC3) that were specifically occupied in the SCLC-Y subtype and expressed at higher levels in the SCLC-Y subtype than in the SCLC-A subtype. Hippo pathway-related genes, such as YAP1, WW domain containing transcription regulator 1, large tumor suppressor kinase 2, and REST, which were not contained in the SCLC-A subtype, were identified in this category (Fig. 1b, Supplementary Table S1). To validate functional differences in SOX2 between the SCLC-A and SCLC-Y subtype SCLC, we used the DAVID online bioinformatics resources 6.7 for GO functional analysis and extracted the top ten enriched categories in biological processes. Neuron-related categories were more likely to be included in the SCLC-A subtype, and cell development- or movement process-related categories in the SCLC-Y subtype (Fig. 1c and Supplementary Table S1). These results suggest that SOX2 regulates distinct transcriptional programs between the SCLC-A and SCLC-Y subtype cell lines.
ASCL1 is one of the driver molecules of SOX2 and recruits SOX2 for distinct transcriptional regulation in a SCLC-A subtype cell line
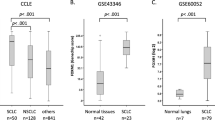
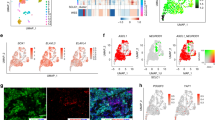
To investigate the biological effects of ASCL1 on SOX2 in lung cancer cell lines, we performed ASCL1 transfection experiments. An A549 cell line that stably expresses ASCL1 was established, as previously reported [17, 20, 21]. A549 is a representative ADC cell line that does not express ASCL1 or other NE markers. A549 expressed SOX2 more weakly than the SCLC cell lines examined (Fig. 1a). We detected upregulated expression of SOX2 following the forced expression of ASCL1 in A549 cells. The expression of INSM1 and WNT11 was also induced (Fig. 2a). IHC staining revealed that SOX2 was more strongly expressed in xenotransplanted tumor cells with the ASCL1 transgene than in those from A549 mock cells (Fig. 2b). To show changes in transcriptional regulation driven by SOX2 in an ASCL1-dependent manner, we compared the target genes of SOX2 between ASCL1-transfected A549 and A549 mock cells. We combined the results of ChIP- and RNA-seq analyses for these cells and identified 35 genes that had specific SOX2-binding peaks and higher expression levels of mRNAs in ASCL1-transfected A549 cells than in mock cells (Fig. 2c). Five genes, INSM1, SV2B, adenosine A2a receptor (ADORA2A), OUT deubiquitinase with linear linkage specificity like (OTULINL), and ArfGAP with coiled-coil, Ankyrin repeat and PH domains 3 (ACAP3), were shared with 346 SOX2-bound genes that were specific for SCLC-A subtype cell lines. These results suggested that SOX2 drives the distinct transcriptional regulation under the presence of ASCL1 in this cell. An IGV snapshot showed SOX2 binding at the overlapping region of the TSS of INSM1 and ASCL1 in SCLC-A subtype cells H69 and H889. After the transfection of ASCL1, it was clearly shown that SOX2 recruitment starts at the overlapping region of TSS of INSM1 in A549 cells (Fig. 2d). To confirm that ASCL1 actually affects SOX2 expression in SCLCs, we conducted ASCL1 knockdown experiments using RNAi in H69, H889, and SBC1 cell lines as representatives of SCLC cells that simultaneously express ASCL1 and SOX2. The knockdown of ASCL1 expression in these cells resulted in significant reductions in SOX2 in two out of the three cell lines, namely, H889 and SBC1 (Fig. 2e). Furthermore, to examine SOX2, ASCL1, and INSM1 expression patterns, we stained 30 surgically resected SCLC tissues for these proteins as well as 20 surgically resected ADC and 20 surgically resected SCC tissues for SOX2 and ASCL1. IHC revealed that SCLCs and SCCs expressed SOX2 at slightly higher levels than ADCs: ~70.0% in SCLCs, 55.0% in SCCs, and 35.0% in ADCs. In SCLC tissues, 60% of cases (18 out of 30 cases) were double positive for ASCL1 and SOX2 and were also positive for INSM1. Although SOX2-positive, ASCL1-negative cases were detected (3 out of 30 cases), there were no ASCL1-positive, SOX2-negative cases (0 out of 30 cases). INSM1 was strongly expressed in most SCLC cases (25 out of 30 cases) and all ASCL1-positive cases simultaneously expressed INSM1 (18 out of 18 cases) (Fig. 2f, Table 3, and Supplementary Fig. S3). Furthermore, based on the results showing that SOX2, ASCL1, and INSM1 were more likely to be coexpressed in SCLC, we surveyed public datasets of gene expression profiling in human SCLC samples and examined their relationships. The RNA-seq dataset using tumor samples from 79 SCLC patients confirmed the coordinated expression of ASCL1 and SOX2 in human SCLC tissue samples (GSE60052: ρ = 0.327759, p = 0.003168). In the same manner, we confirmed the coordinated expression of ASCL1 and INSM1 (GSE60052: ρ = 0.357944, p = 0.001188). Heatmap data focusing on SOX2, ASCL1, and INSM1 were obtained using the dataset reported by Jiang et al. [22] (Fig. 2g). We obtained similar results using the other public datasets (GSE62021 [23], GSE36139 [24], and the paper-accompanied data from George et al. (NIHMS782739) [25]) (Supplementary Fig. S4). These results support the positive regulation of SOX2 and INSM1 expression by ASCL1.
a WB analysis showed that the transfection of ASCL1 in A549 ADC cells increased SOX2, INSM1, and WNT11 expression. β-ACTIN served as an internal control. b Tumor tissues by the xenotransplantation of mock A549 cells and ASCL1-transfected A549 cells in immunodeficient mice. Using immunohistochemistry (IHC), ASCL1 transfection induced SOX2 protein expression in tumor cell nuclei. Representative images are shown. Scale bar = 200 μm. c Changes in transcriptional regulation driven by SOX2 between ASCL1-transfected A549 and A549 mock cells. ChIP- and RNA-seq combined data showed 35 specific SOX2-bound genes in ASCL1-transfected A549. INSM1 was newly detected after the transfection of ASCL1. Five genes, INSM1, SV2B, ADORA2A, OTULINL, and ACAP3, were shared with 346 SOX2-bound genes that were specific for SCLC-A subtype cell lines (red color). d An integrated genome viewer snapshot showed SOX2 binding at the overlapping region of the transcription starting site of the INSM1 and ASCL1 gene in SCLC cell lines. The fold enrichment values of SOX2 peak on INSM1 loci calculated by MACS2 are 23.79 in H69, 19.1 in H889, 8.79 in A549 ASCL1-TF, and no enrichment in SBC3 and A549 cells. The fold enrichment value on ASCL1 loci is 7.17 in H889 cells. These values are calculated against random Poisson distribution with local lambda. e The suppression of SOX2 by RNAi for ASCL1 was confirmed in H889 and SBC1 cells by WB analysis. f IHC images of surgically resected SCLC tissues for SOX2, ASCL1, and INSM1. These proteins were strongly expressed in tumor cell nuclei. Representative images are shown. Scale bar = 200 μm. g Expression levels of ASCL1, INSM1, and SOX2 in the RNA-seq dataset of SCLC tissues. The GSE60052 (n = 79) dataset [20] was analyzed. NT nontreated, si small interfering.
The role of SOX2 in the SCLC-A subtype cell lines and the ASCL1-transfected A549 cell line
To investigate the biological significance of SOX2 in the SCLC-A subtype cell lines and the ASCL1-transfected A549 cell line, we conducted SOX2 knockdown experiments using RNAi on these cells. The knockdown of SOX2 expression resulted in significant reductions in ASCL1 and INSM1 expression in the H69, H889, and SBC1 cell lines (Fig. 3a). Furthermore, we previously reported that ASCL1 regulates WNT11 and CDH1 expression [17, 21]. The expression of WNT11 and CDH1 was reduced in the H69, H889, and SBC1 cell lines after the knockdown of SOX2 (Fig. 3a). This result suggests that SOX2 affected the expression of EMT-markers in SCLC. The results of the SOX2 knockdown experiment on ASCL1-transfected A549 cells are also shown in Fig. 3. Not only INSM1 and WNT11, but also NOTCH1, MYC, TCF4, RBL1, and Trp53 protein levels decreased after the knockdown of SOX2. The suppression of SOX2 also affected the phosphorylation of histone H3 (p-HH3) protein levels in SCLC-A subtype cell lines and the ASCL1-transfected A549 cell line (Fig. 3b). Cell counting assays revealed that the knockdown of SOX2 suppressed cell growth in the H69, H889, and SBC1 cells lines (Fig. 3c). This result suggests that SOX2 positively affected cell proliferation in the SCLC-A subtype cell lines and regulated the expression of key molecules for the SCLC phenotype in the presence of ASCL1. We also assessed the changes of mRNA expression levels corresponding to these proteins as being altered following SOX2 knockdown in each cell line. While most of the genes behaved in concordance with the altered patterns of protein expression, a few genes, such as INSM1 in SBC1 cells or NOTCH1 and TP53 in ASCL1-transfected A549 cells, did not have altered mRNA expression levels after the knockdown of SOX2. This may suggest that SOX2 regulates not only molecular transcription, but also protein stability or degradation in a cell context-dependent manner.
a SOX2 affects INSM1, ASCL1, WNT11, and CDH1 expression in SCLC cell lines. The suppression of INSM1, ASCL1, WNT11, CDH1, and p-HH3 was observed in the SCLC-A subtype cell lines (H69, H889, and SBC1s) with RNAi for SOX2. b SOX2 was involved in the expression of NOTCH1, MYC, TCF4, Trp53, and RBL1 in ASCL1-TF A549 cells. We also evaluated the changes of mRNA expression levels corresponding to the proteins that were altered following SOX2 knockdown in each cell line by qRT-PCR. c Cell counting assays with SCLC cell lines. The cell growth curve is shown. The suppression of cell proliferation was observed in H69, H889, and SBC1 cells with RNAi against SOX2. The analysis was performed in triplicate. Data are shown as the mean ± SD. Asterisks indicate a significant difference; *p < 0.05.
The role of SOX2 in SCLC-Y subtype cell lines
To investigate the role of SOX2 in SCLC-Y subtype cell lines, we conducted SOX2 knockdown experiments using RNAi on SBC3 and H69AR cell lines. SBC3 and H69AR are representative SCLC-Y subtype cell lines that lack the expression of ASCL1 and INSM1, but increase the expression of YAP1 (Fig. 1a). These cells have markedly fewer NE properties than the SCLC-A subtype cell lines [20]. WB analysis revealed that Hippo-related molecules, such as YAP1 or TEAD1, and VIMENTIN expression levels were significantly reduced after the knockdown of SOX2. qRT-PCR also displayed similar results. On the other hand, the expression of ASCL1, INSM1, and WNT11 was not significantly affected by the knockdown of SOX2 in these cells (Fig. 4a). Furthermore, SOX2 knockdown did not significantly reduce tumor cell proliferative capacity in the SBC3 and H69AR cell lines (Fig. 4b). These results suggest that SOX2 did not always sufficiently affect tumor proliferative capacity in SCLC and, in the SCLC-Y subtype, it regulated the expression of downstream target genes that were distinct from those of the SCLC-A subtype. We obtained similar results in the SOX2 knockout experiments using the CRISPR/Cas9 system in the SBC3 and H69AR cell lines. qRT-PCR revealed that Hippo-related molecules, such as YAP1 or TEAD1, and VIMENTIN expression levels were also significantly reduced after the knockout of SOX2 in the SCLC-Y subtype. In addition, the knockout of SOX2 in the SCLC-Y subtype also did not significantly reduce tumor cell proliferative capacity in vitro and ex vivo (Supplementary Fig. S5).
a YAP1, TEAD1, or VIMENTIN protein and mRNA expressions decreased in SBC3 and H69AR cells with RNAi against SOX2. ASCL1, INSM1, and WNT11 mRNA expression did not change in these cell lines. b SOX2 protein and mRNA expression significantly decreased, whereas tumor proliferative capacity did not decrease after SOX2 knockdown in SBC3 and H69AR cells. The analysis was performed in triplicate. Data are shown as the mean ± SD. Asterisks indicate a significant difference; *p < 0.05.
Discussion
SOX2 is an oncogene in human SCLC and its amplification has been detected in some SCLCs. Sholl et al. reported that 72% of high-grade NE carcinomas were positive for SOX2 by immunohistochemistry [26], and we validated a similar frequency (70%) in our SCLC tissue samples. In the present study, we focused on the role of SOX2 in SCLC. Furthermore, we investigated the different functions of SOX2 and whether ASCL1 was present in SCLC cell lines. SOX2 regulated INSM1 or WNT11 with the cooperation of ASCL1 in the SCLC-A subtype. On the other hand, in the SCLC-Y subtype, SOX2-targeted distinct genes such as those in the Hippo pathway. These results suggest that SOX2 is not only an oncogene in human SCLC, but may also drive distinct transcriptional regulation in each subtype of SCLC. Therefore, care is needed when considering SOX2 as a potential therapeutic target or biological marker in the diagnosis and treatment of SCLC.
Historically, Gazdar et al. reported the different forms of the “classic” and “variant” subtype of SCLC. Classical SCLC cells are characterized by floating cell growth and distinct NE differentiation, as well as variant SCLC cells by adhesive growth and poor NE differentiation [27, 28]. It may be possible that SCLC-A subtype belongs to the classical subtype and SCLC-Y subtype belongs to the variant subtype of SCLC. Borromeo et al. reported that Ascl1 played a pivotal role in tumorigenesis in mouse models of SCLC, and also suggested that SOX2 and INSM1 were target genes of ASCL1 [9]. Osada et al. reported roles for ASCL1 in CDH1 expression and NE differentiation. Furthermore, the knockdown of ASCL1 induced growth inhibition and apoptosis in SCLC cell lines [29, 30]. INSM1 is a crucial regulator of NE differentiation in lung cancer, and is specifically expressed in SCLC, along with ASCL1 [19]. Our results support these findings, and, as a novel insight, ASCL1 and SOX2 cooperatively modulated NE differentiation or EMT-marker expression in human SCLC. The knockdown of SOX2 expression in the examined SCLC-A subtype cell lines resulted in significant reductions in WNT11 and CDH1. We previously showed that ASCL1 induced the EMT-like phenotype in A549 ADC cells [21] and demonstrated that WNT11 regulated CDH1 expression in SCLC cell lines [17]. SOX2 may potently modulate NE differentiation and CDH1 expression via ASCL1 or WNT11 in SCLC.
The results of molecular analysis of SOX2 in ASCL1-transfected A549 cells are of interest. After the overexpression of ASCL1, SOX2 expression was enhanced and INSM1 was simultaneously expressed in the cells. The combination analysis of ChIP- and RNA-seq identified 35 genes, including INSM1, that had specific SOX2-binding peaks and higher expression levels of mRNAs in ASCL1-transfected A549 cells than in mock cells. The knockdown of SOX2 in ASCL1-transfected cells caused the suppression of INSM1, suggesting that ASCL1 activates SOX2 and regulates INSM1 expression together with SOX2. More interestingly, while the knockdown of ASCL1 using RNAi decreases SOX2 in two of the three SCLC-A subtype cell lines, H889 and SBC1 (Fig. 2e), the knockdown of SOX2 in each of these three SCLC-A subtype cell lines, H69, H889, and SBC1, decreased expression of ASCL1 (Fig. 3a). This suggests that a positive regulatory loop could exist between ASCL1 and SOX2 in some SCLC-A subtype cells. Contrary to the results of H889 and SBC1 cells, there would be exceptions, such as H69 cell, which we could not demonstrate the direct relationships for protein expression between ASCL1 and SOX2 (Fig. 2e), or INSM1 (data not shown). This might suggest that not all of ASCL1 in SCLC-A subtype cells could recruit SOX2 to the INSM1 gene. But, it is quite possible that ASCL1 is needed for the recruitment of SOX2 to the INSM1 gene, as shown in our ASCL1 transgene or RNAi experiments. Zhang et al. reported that SCLC specifically upregulated a series of TFs such as ASCL1 or INSM1 [31]. Though we have compared our analysis result with their work, we could not detect other intriguing genes except for ASCL1 and INSM1. But, in the present study, we newly demonstrated the mechanism of INSM1 or ASCL1 gene transcriptional regulation via SOX2 in SCLC-A subtype. In addition, we previously reported that ASCL1 regulates WNT11 gene expression [17]. We speculate that a regulatory mechanism of WNT11 gene expression occurs via SOX2 in the presence of ASCL1. Furthermore, NOTCH1, TCF4, MYC, Trp53, and RBL1 expression decreased after the knockdown of SOX2 in these cells. Intratumoral heterogeneity generated by Notch signaling has been shown to promote SCLC [6]. The Notch1–Hes1 pathway is a repressor of NE differentiation through the decreased expression of NE-promoting TFs such as ASCL1 and INSM1 [19, 32, 33]. Chen et al. reported that silencing of the SOX2 gene reduced the tumorigenic properties of A549 cells with the attenuated expression of MYC and NOTCH1 in xenografted tumors in NOD/SCID mouse [34]. The canonical Wnt pathway induces MYC through TCF4 and other Wnt signal components. In addition, WNT11 has been reported to activate both canonical and non-canonical Wnt pathways [35, 36]. Moreover, mouse models carrying conditional alleles for both Trp53 and Rb1 developed small cell carcinoma in the lung [37]. The universal bi-allelic inactivation of Trp53 and RB1 was previously reported in human samples [25]. Meder et al. showed that NOTCH, ASCL1, Trp53, and RB alterations defined an alternative pathway driving NE and small cell carcinomas [38]. Moreover, the ablation of all three retinoblastoma family members, Rb1, Rbl1, and Rbl2, in the mouse lung resulted in the formation of NE tumors [39]. SOX2 has been suggested to play an important role, particularly in an ASCL1-dependent manner, in the SCLC phenotype and tumorigenesis in the interaction with these principle tumor suppressants.
We performed SOX2 knockdown or knockout experiments using RNAi or the CRISPR/Cas9 system in the SCLC-Y subtype, SBC3, and H69AR cells. After SOX2 knockdown or knockout, YAP1, TEAD1, and VIMENTIN protein and mRNA expression levels decreased in these cells. A previous study reported that YAP1, which is the main Hippo pathway effector, was frequently lost in high-grade NE lung tumors, and showed reciprocal expression against INSM1 [40]. Among SCLC, the loss of YAP1 correlated with the expression of NE markers, and survival analysis revealed that YAP1-negative cases were more chemo-sensitive than YAP1-positive cases [41]. The YAP/TAZ subgroup displayed an adherent cell morphology [42] and lower expression levels of ASCL1. REST was also included in the genes of SOX2-bound sites in the SCLC-Y subtype. REST encodes a transcriptional repressor that represses neuronal and NE genes in nonneuronal and non-NE tissues and, thus, serves as a negative regulator of neurogenesis including SCLC [7, 43, 44]. We demonstrated that SOX2 modulated the Hippo pathway in the SCLC-Y subtype cell lines in the present study. In the SCLC-A subtype cell lines, CDH1 expression decreased after the knockdown of SOX2 by RNAi (Fig. 3a). These results suggest that SOX2 potently modulated EMT-marker expression in lung cancer via the Hippo or Wnt signaling pathway in SCLC.
Rudin et al. reported that the knockdown of SOX2 by doxycycline-inducible shRNA inhibited cell proliferation in SCLC cell lines [10]. In the SCLC-A subtype cell lines, we demonstrated that SOX2 knockdown using RNAi decreased tumor cell proliferative capacity. In contrast, the knockdown of SOX2 in the SCLC-Y subtype cells did not significantly affect their cell proliferative capacity. Furthermore, while we failed to establish SOX2-knockout SCLC-A subtype cell lines because of high lethality (data not shown), we successfully established SOX2-knockout SCLC-Y subtype cell lines with sufficient proliferative capacity (Supplementary Fig. S5). This functional discrepancy may be attributed to functional differences in SOX2 between the SCLC-A and SCLC-Y subtype cell lines. Currently, SCLC is divided into four subtypes according to the dominant expression of TFs that are considered as the master regulators of SCLC: ASCL1, NEUROD1, YAP1, and POU2F3 [4]. In the present study, we revealed that SOX2 cooperated with the key regulatory molecules, such as ASCL1 and YAP1, in SCLC, and showed that SCLC may be divided into three groups based on the expression of ASCL1 and SOX2: ASCL1–SOX2 double high, ASCL1 low and SOX2 high, and ASCL1–SOX2 double low (Fig. 2f, g). In addition to genomic profiling, which has been adopted in clinical practice, several research initiatives that catalog DNA, RNA, and protein profiles among lung SCC and ADC have accelerated the pace of discovery, such as The Cancer Genome Atlas. However, similar efforts have not yet been achieved for SCLC due to the lack of adequate tumor tissues [2]. This needs to be investigated in a large prospective or cohort study in the future.
In summary, the SCLC-A subtype frequently and strongly expresses both SOX2 and ASCL1. ASCL1-recruited SOX2 plays an important role in driving distinct transcriptional regulation. We demonstrated that SOX2 regulates lineage-specific genes, such as INSM1, in the SCLC-A subtype. While we revealed the importance of SOX2 for cell growth and the modulation of EMT-marker expression, we detected a functional discrepancy in SOX2 between the SCLC-A and SCLC-Y subtype (Fig. 5). The present results suggest that the ASCL1–SOX2 axis is extremely important as a potential therapeutic target or biological marker in the SCLC-A subtype. On the other hand, the fundamental role of SOX2 in the SCLC-Y subtype, in which ASCL1 is negative, was suggested by the activation of the Hippo signaling pathway. Further studies on SOX2 that focuses on highly specific molecules, such as those involved in the recruitment of SOX2 in the other SCLC subtypes, are needed. The present study promotes our understanding of the significance of SOX2 in SCLC, which will hopefully lead to the development of novel targeted therapies and better prognoses for patients with SCLC.
ASCL1 and SOX2 cooperatively regulate INSM1 and WNT11 expression and SOX2 affects not only tumor cell proliferation, but also modulation of EMT-marker expression or NE differentiation in the SCLC-A subtype. On the other hand, SOX2 affects distinct cell signaling, such as the Hippo pathway, and modulates EMT-marker expression in the SCLC-Y subtype. SOX2 affects tumor cell survival slightly more in the SCLC-A subtype than in the SCLC-Y subtype cells.
Dataset availability
References
Rodriguez E, Lilenbaum RC. Small cell lung cancer: past, present, and future. Current Oncol Rep. 2010;12:327–34.
Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer. 2015;121:664–72.
Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17:765.
Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97.
Ball DW, Azzoli CG, Baylin SB, Chi D, Dou S, Donis-Keller H, et al. Identification of a human achaete-scute homolog highly expressed in neuroendocrine tumors. Proc Natl Acad Sci USA. 1993;90:5648–52.
Lim JS, Ibaseta A, Fischer MM, Cancilla B, O’Young G, Cristea S, et al. Intratumoural heterogeneity generated by Notch signaling promotes small-cell lung cancer. Nature. 2017;545:360–4.
Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res. 2018;7:32–49.
Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci USA. 2014;111:14788–93.
Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–72.
Rudin CM, Durinck S, Atawiski F, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6.
Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. WHO classification of tumours of the lung, pleura, thymus and heart. 4th ed. Lyon, France: IARC; 2015. p. 63–8.
Yoshida R, Nagata M, Nakayama H, Niimori-Kita K, Hassan W, Tanaka T, et al. The pathological significance of Notch1 in oral squamous cell carcinoma. Lab Investig. 2013;93:1068–81.
Adachi K, Nikaido I, Ohta H, Ohtsuka S, Ura H, Kadota M, et al. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol Cell. 2013;52:380–92.
Niwa H, Nakamura A, Urata M, Shirae-Kurabayashi M, Kuraku S, Russell S, et al. The evolutionally-conserved function of group B1 Sox family members confers the unique role of Sox2 in mouse ES cells. BMC Evol Biol. 2016;16:173.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4:44–57.
Tenjin Y, Kudo S, Kubota S, Yamada T, Matsuo A, Sato Y, et al. Ascl1-induced Wnt11 regulates neuroendocrine differentiation, proliferation and E-cadherin expression in small-cell lung cancer and Wnt11 regulates small-cell lung cancer biology. Lab Investig. 2019;99:1622–35.
Ran F, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Fujino K, Motooka Y, Hassan WA, Ali Abdalla MO, Sato Y, Kudoh S, et al. INSM1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol. 2015;185:3164–77.
Kudoh S, Tenjin Y, Kameyama H, Ichimura T, Yamada T, Matsuo A, et al. Significance of achaete-scute complex homologue 1 (ASCL1) in pulmonary neuroendocrine carcinomas; rna sequence analyses using small cell lung cancer cells and ascl1-induced pulmonary neuroendocrine carcinoma cells. Histochem Cell Biol. 2020. https://doi.org/10.1007/s00418-020-01863-z.
Ito T, Kudoh S, Ichimura T, Fujino K, Hassan WA, Udaka N. Small cell lung cancer, an epithelial to mesenchymal transition (EMT)-like cancer: significance of inactive Notch signaling and expression of achaete-scute complex homologue 1. Hum Cell. 2017;30:1–10.
Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z, Yao X, et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genet. 2016;12:e1005895.
Saito Y, Nagae G, Motoi N, Miyauchi E, Ninomiya H, Uehara H, et al. Prognostic significance of CpG island methylator phenotype in surgically resected small cell lung carcinoma. Cancer Sci. 2016;107:320–5.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53.
Sholl LM, Long KB, Hornick JL. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol. 2010;18:55–61.
Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res. 1985;45:2924–30.
Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–23.
Osada H, Tatematsu Y, Yatabe Y, Horio Y, Takahashi T. ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res. 2005;65:10680–5.
Osada H, Tomida S, Yatabe Y, Tatematsu Y, Takeuchi T, Murakami H, et al. Roles of achaete-scute homologue 1 in DKK1 and E-cadherin repression and neuroendocrine differentiation in lung cancer. Cancer Res. 2008;68:1647–55.
Zhang S, Li M, Ji H, Fang Z. Landscape of transcriptional deregulation in lung cancer. BMC Genom. 2018;19:435.
Ball DW. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett. 2004;204:159–69.
Hassan WA, Yoshida R, Kudoh S, Hasegawa K, Niimori-Kita K, Ito T. Notch1 controls cell invasion and metastasis in small cell lung carcinoma cell lines. Lung Cancer. 2014;86:304–10.
Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L, et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS ONE. 2012;7:e36326.
Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356.
Rapp J, Jaromi L, Kvell K, Miskei G, Pongracz JE. WNT signaling—lung cancer is no exception. Respir Res. 2017;18:167.
Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Introduction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9.
Meder L, König K, Ozretić L, Schultheis AM, Ueckeroth F, Ade CP, et al. NOTCH, ASCL1, p53, and RB alterations define an alternative pathway driving neuroendocrine and small cell carcinomas. Int J Cancer. 2016;138:927–38.
Lázaro S, Pérez-Crespo M, Enguita AB, Hernández P, Martínez-Palacio J, Oteo M, et al. Ablating all three retinoblastoma family members in mouse lung leads to neuroendocrine tumor formation. Oncotarget. 2017;8:4373–86.
McColl K, Wildey G, Sakre N, Lipka MB, Behtaj M, Kresak A, et al. Reciprocal expression of INSM1 and YAP1 defines subgroups in small cell lung cancer. Oncotarget. 2017;8:73745–56.
Ito T, Matsubara D, Tanaka I, Makiya K, Tanei ZI, Kumagai Y, et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci. 2016;107:1527–38.
Horie M, Saito A, Ohshima M, Suzuki HI, Nagase T. YAP and TAZ modulate cell phenotype in a subset of small cell lung cancer. Cancer Sci. 2016;107:1755–66.
Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cell. J Neurosci. 2011;31:9772–86.
Thiel G, Ekici M, Rossler OG. RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res. 2015;359:99–109.
Acknowledgements
We thank Ms. Yuko Fukuchi and Ms. Takako Maeda for their technical assistance, the staff of LILA for their technical support, the Institute of Molecular Embryology and Genetics, Kumamoto University, for help with RNA-seq analysis. This study was partially supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (numbers 18K19480 and 20H03691), and by Aihara Pediatric and Allergy Clinic, Yokohama, Japan. This study was also supported in part by the program of the Joint Usage/Research Center for Developmental Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tenjin, Y., Matsuura, K., Kudoh, S. et al. Distinct transcriptional programs of SOX2 in different types of small cell lung cancers. Lab Invest 100, 1575–1588 (2020). https://doi.org/10.1038/s41374-020-00479-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-00479-0
This article is cited by
-
WNT ligands in non-small cell lung cancer: from pathogenesis to clinical practice
Discover Oncology (2023)
-
DNA-methylation patterns imply a common cellular origin of virus- and UV-associated Merkel cell carcinoma
Oncogene (2022)
-
ASCL1 activates neuronal stem cell-like lineage programming through remodeling of the chromatin landscape in prostate cancer
Nature Communications (2022)
-
Insulinoma-associated-1 (INSM1) expression in thymic squamous cell carcinoma
Virchows Archiv (2022)