Abstract
Fibroblast-like synoviocytes (FLS) are major contributors to joint inflammation in rheumatoid arthritis (RA). Forkhead box O 3 (FOXO3) perturbations in immune cells are increasingly linked to RA pathogenesis. Here, we show that FOXO3 is distinctly inactivated/phosphorylated in the FLS of rheumatoid synovitis. In vitro, stimulation of FLS with tumor necrosis factor-alpha α (TNFα) induced a rapid and sustained inactivation of FOXO3. mRNA profiling revealed that the inactivation of FOXO3 is important for the sustained pro-inflammatory interferon response to TNFα (CXCL9, CXCL10, CXCL11, and TNFSF18). Mechanistically, our studies demonstrate that the inactivation of FOXO3 results from TNF-induced downregulation of phosphoinositide-3-kinase-interacting protein 1 (PIK3IP1). Thus, we identified FOXO3 and its modulator PIK3IP1 as a critical regulatory circuit for the inflammatory response of the resident mesenchymal cells to TNFα and contribute insight into how the synovial tissue brings about chronic inflammation that is driven by TNFα.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease. Uncontrolled persistent inflammation of the synovial membrane leads to progressive joint damage and disability [1]. Reciprocal crosstalk between infiltrating immune cells and resident fibroblasts (fibroblast-like synoviocytes; FLS) are thought to amplify and perpetuate joint inflammation [2]. In response to immune cell derived pro-inflammatory mediators (e.g. tumor necrosis factor-alpha; TNFα) FLS secrete large amounts of cytokines (e.g. Interleukin-6 (IL6)) and chemokines (e.g. CXC-Ligand (CXCL)-9, CXCL10) [3], which further enhance immune cell recruitment and activation, thereby promoting synovial inflammation [2]. Interrupting this vicious circle by targeting FLS has therefore emerged as a potential therapeutic strategy for RA. For this, understanding of how FLS respond to and shape the synovial, inflammatory microenvironment is required. Signaling circuits that activate FLS in the context of joint inflammation are emerging. Among others, the phosphatidylinositol-3-kinase/Protein kinase B (PI3K/AKT) pathway directs the inflammatory behavior of FLS [4,5,6]. In response to extracellular signals such as growth factors or inflammatory mediators PI3K promotes the activation of AKT [7, 8]. Once activated, AKT phosphorylates and distinctly controls downstream effectors, such as the mechanistic target of rapamycin (mTOR) and the forkhead box O (FOXO) transcription factors [8]. In arthritis, persistent activation of the PI3K/AKT pathway was observed in both animal models and human synovial tissues [9, 10]. Blocking PI3K by genetic or pharmacological approaches ameliorated joint inflammation in various animal models [5, 11, 12]. Importantly, this pathway operates in FLS and might thereby contribute to joint inflammation [4,5,6]. The effectors downstream of PI3K/AKT in FLS, however, remain to be elucidated.
Here, we explore the role of the transcription factor FOXO3 [13] in FLS inflammation. Recently, FOXO3 has received attention since GWAS data linked FOXO3 activity with RA. A single-nucleotide polymorphism (SNP) in FOXO3 (rs12212067; T > G) was associated with lower RA activity scores and less radiographic progression [14, 15]. Correspondingly, collagen-induced arthritis (CIA) was pronounced in mice with loss-of-FOXO3 function [14]. By contrast, FOXO3 deficiency ameliorated symptoms in the K/BxN serum transfer arthritis model. In this model, FOXO3 was shown to support neutrophil survival during inflammation [16]. This conflicting data suggest that FOXO3 may exhibit a cell-type specific pro- or anti-inflammatory role. Nonetheless FOXO3 likely mediates critical fate decisions in synovial inflammation. To unravel its potential pathogenic involvement in RA, we explore the biological consequences of FOXO3 regulation in FLS.
Material and methods
Immunohistochemistry
Synovial biopsy specimens were obtained from RA (fulfilling ACR/EULAR classification criteria for RA [17]) and OA patients at the time of joint replacement or synovectomy (patient characteristics are shown in Supplementary Table 1). After fixation with paraformaldehyde, synovial tissues were embedded in paraffin. Paraffin-embedded sections were treated with Tris-EDTA (pH 9) and then incubated with goat serum. p-FOXO3 was detected by incubating sections with a polyclonal rabbit anti-p-FOXO3 antibody (Abcam). A non-immune immunoglobulin of the same isotype and concentration as the primary antibody (anti-rabbit IgG (R&D) was used as a control. After incubation with a biotinylated goat anti-rabbit antibody (Vector), sections were incubated with Vectastain Elite reagent and visualized using 3,3-diaminobenzidine (Vector). Sections were counterstained with hematoxylin (Merck). Pictures were taken with an Axioskop 2 microscope (Zeiss) equipped with a digital camera (Olympus). Expression of p-FOXO3 was determined using semi-quantitative scoring (0 = no staining, 3 = high staining).
Isolation and culture of RA-FLS
FLS were obtained from discarded synovial tissues from RA patients (fulfilling ACR/EULAR classification criteria for RA [17]) undergoing joint replacement or synovectomy. FLS were isolated as previously described [18]. FLS were cultured in DMEM (GIBCO by life technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone), 1% penicillin/streptomycin (P/S), and nonessential amino acids (both GIBCO). FLS between passages 4 and 8 were used for all experiments.
Cytokines and inhibitors
Recombinant human TNFα was purchased from R&D. The PI3K-inhibitors, Duvelisib (IPI-145) and Idelalisib (CAL-101), the AKT-inhibitor Ipatasertib (GDC-0068) and the mTOR-inhibitor Torin-1 were obtained from Selleckchem.
Western blots
RA-FLS were lysed with RIPA-buffer (Thermo Fisher) supplemented with Halt™ phosphatase inhibitor cocktail (Thermo Fisher) and protease inhibitor mix (Sigma Aldrich). Protein lysates were fractioned on polyacrylamide gels followed by electro transfer to nitrocellulose membranes, which were blocked with either 5% BSA or 5% non-fat dry milk and then incubated with primary antibodies (Cell Signaling: p-FOXO3 (T32), FOXO3, p-S6K (T389); Santa Cruz: S6K; Protein Tech: PIK3IP1; Sigma Aldrich: ACTIN). After incubation with HRP-conjugated secondary antibodies (Cell Signalling) specific bands were detected with the BIORAD Clarity ECL Western substrate. Reblots were performed using ReBlot Plus Strong Solution (Merck).
Real-time quantitative reverse transcription PCR
RNA was isolated with the RNeasy Mini Kit (Qiagen) and then reverse transcribed into cDNA with the Omniscript RT kit (Qiagen). RNA concentration was determined using a Nanodrop spectrophotometer. Quantitative PCR (qPCR) was performed using the Fast Start SYBR Green I kit (Roche) and the Roche Light Cycler. Results were quantified by the 2−ΔΔC(t) method, using GAPDH expression levels for normalization. Primer sequences: CXCL9 Forward: ATCAGCACCAACCAAGGGACT, Reverse: GCTTTTTCTTTTGGCTGACCTG; CXCL10 Forward: ATTTGCTGCCTTATCTTTCTG, Reverse: TCTCACCCTTCTTTTTCATTGTAG; CXCL11 Forward: GAAGGATGAAAGGTGGGTGA, Reverse: AAGCACTTTGTAAACTCCGATG; PIK3IP1 Forward: CCAGTGATTGGGATCAGCCA, Reverse: TCCCCCTCTTGTAGGAGTAGC; TNFSF18 Forward: ACGCAAGGAGGTTCAGAAGA, Reverse TCTTTGCTCCTTCAGTTGGC; GAPDH Forward: TGATGACATCAAGAAGGTGGTGAAG, Reverse TCCTTGGAGGCCATGTGGGCCAT;
ELISA
IL6, IL8, and CXCL10 ELISA kits were purchased from eBioscience and the MMP3 ELISA kit from R&D. ELISA were performed according to the manufacturer´s protocol.
Immunofluorescence staining and automated imaging
RA-FLS were cultured on CellCarrier-96 Ultra Microplates (PerkinElmer). FLS were fixed with 4% paraformaldehyde containing 0,02% Tween-20 and permeabilized with 0.3% Triton X-100. After blocking with 5% bovine serum albumin (BSA), FLS were stained with the primary antibody (monoclonal rabbit anti-FOXO3, Cell Signalling) over night at 4 °C. After incubation with an Alexa Fluor 555 coupled secondary antibody (goat anti-rabbit, Invitrogen) nuclei were counterstained with DAPI (Sigma Aldrich). Pictures were taken with the Opera Phenix high-content screening system (PerkinElmer) at ×20 magnification. Images from automated imaging were analyzed with CellProfiler v3.0.0 (http://cellprofiler.org/citations/). DAPI signal was used to define the nuclear area. Single cells were selected by a neighborhood-distance of more than 12 pixel (7 µm) to avoid overlapping areas. The perinuclear region was defined as a 10 pixel (5 µm) wide ring around the nucleus. FOXO3 signal was quantified in both regions and log2 ratios of nuclear to perinuclear mean intensities were calculated for every individual cell. Plotting and statistical analysis were performed in R (version 3.4.3). Representative pictures were taken with a Leica TCS SP5 (Leica Microsystems) or an Olympus IX83 microscope.
siRNA-mediated knockdown
For siRNA-mediated knockdown RA-FLS were cultured in Opti-MEM (GIBCO). siRNA pools (ON-TARGETplus: FOXO3: L-003007-00-0005, PIK3IP1: L-003007-00-0005, Dhamarcon) were transfected using Lipofectamine (Thermo Fisher) as described elsewhere [19].
RNA sequencing
RNA was isolated using the RNeasy purification kit (Quiagen). RNA-seq libraries were prepared with TruSeq stranded mRNA sample preparation kit (Illumina) using Sciclone and Zephyr liquid handling robotics (PerkinElmer). The libraries were sequenced on HiSeq 3000 (Illumina) platform using 50 bp single-read chemistry.
Processing of RNA sequencing data
Raw sequencing data were processed with Illumina2bam (http://github.com/wtsi-npg/illumina2bam) to generate unaligned BAM files. Sequence reads were mapped onto the human genome annotation release hg38 (GRCh38) using STAR aligner (version 2.5.2b) [20]. Reads were counted with the summarizeOverlaps function of Bioconductor package GenomicAlignments (version 1.10.1) [21] on the basis of Ensembl transcript annotation version 89 and DESeq2 (version 1.14.1) [22]. Gene expression values (reads per kilobase exon per million mapped reads (RPKM)) were calculated with Cufflinks (version 2.2.1) [23]. The differential expression between two sample groups was calculated from the raw read counts, with Fisher’s exact test for the significance of all samples together (sum of read counts), and edgeR’s (version 3.12.1) [24] dispersion for the variance within biological replicates. The filtering for differentially expressed genes is for p-value of 0.05 (Bonferroni corrected), maximal dispersion of 0.2 and minimal fold-change of 2. Lists for known PI3K-AKT pathway regulators (GO:0014067; GO:0051898; GO:0014068; GO:0051898) were downloaded from http://amigo.geneontology.org [25, 26]. Heatmap was generated using the pheatmap function in R.
Statistical analysis
We used unpaired and paired t-tests for comparing groups and paired samples provided that the data followed Gaussian distribution. For data that were not normally distributed, the Mann–Whitney U test or Wilcoxon singed-rank test was performed. For all analyses, Graph Pad Prism 6 software was used.
Results
TNF-induced phosphorylation and nucleocytoplasmic shuttling of FOXO3
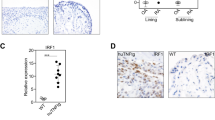
We first examined the degree and pattern of phosphorylated-(p)-FOXO3 in RA synovial tissues. Synovial tissues from osteoarthritis (OA) patients served as non-inflammatory controls. Immunohistochemistry (IHC) revealed significantly increased p-FOXO3 in RA compared to OA synovial tissues (Figs. 1a, b). p-FOXO3 was primarily detected in the hyperplastic synovial lining and in fibroblast-like cells in the synovial sublining layer, suggesting that FOXO3 is phosphorylated in activated RA-FLS.
FOXO3 phosphorylation in the rheumatoid synovium. a Phospho(p)-FOXO3 (S253) expression in paraffin-embedded synovial tissue sections from rheumatoid arthritis (RA) or osteoarthritis (OA) patients (upper panel). Stainings with an isotype matched control antibody (CTRL) are presented in the lower panel. b Synovial tissue sections derived from eleven RA and six OA patients were evaluated for p-FOXO3 expression using semi-quantitative scoring (0 = no staining, 3 = high staining). Wilcoxon–Mann–Whitney test
We have previously shown that TNFα activates AKT [27], which operates upstream of FOXO3 [13]. Therefore we asked whether TNFα might trigger FOXO3 phosphorylation. Indeed, TNFα induced FOXO3 phosphorylation in isolated RA-FLS. TNFα effects were evident after 30 min (Fig. 2a) and sustained throughout 48 hours (Fig. 2b).
TNF-induced nuclear exclusion of FOXO3. a, b Western blots of TNFα treated (10 ng/ml) RA fibroblast-like synoviocytes (RA-FLS). Representative blots of at least eight (a) or three (b) independent experiments with FLS cell lines from different donors. c Boxplots displaying the delta mean of the nuclear to perinuclear FOXO3 signal ratio calculated from automatically captured and analyzed images. FLS were treated with TNFα (10 ng/ml) for the indicated time points. Pooled data from four RA-FLS cell lines from different donors are shown. Unpaired Student’s t-test has been used to assess the statistical significance of untreated vs. treated FLS. *p < 0.05. d Representative pictures of FOXO3 (red) nuclear staining in unstimulated (upper panel) and TNF-stimulated (lower panel) RA-FLS. Actin filaments were labeled with phalloidin (green). Cell nuclei were stained with DAPI (blue)
FOXO3 transcriptional activity is controlled by its retention within or exclusion from the nucleus. FOXO3 phosphorylation facilitates nuclear export, which inhibits its transcriptional function [13]. Since TNF-stimulation results in FOXO3 phosphorylation, we reasoned that TNFα might induce FOXO3 nuclear exclusion. Immunofluorescence (IF) automated microscopy revealed decreased nuclear contents of FOXO3 in TNF-treated FLS (Figs. 2c and 2c). Upon TNFα stimulation FOXO3 was rapidly exported, but returned to the nucleus within 80 min. This was followed by a second wave of FOXO3 nuclear exclusion, which was maintained throughout the timeframe of the experiment.
Our data suggest that FOXO3 is phosphorylated (inactive) in the inflamed rheumatoid synovium. Further, we show that TNFα promotes FOXO3 nuclear export and might thereby alter FOXO3 transcriptional activity.
FOXO3 regulates the TNF-induced interferon response in FLS
FOXO3 acts as transcriptional activator or repressor [28]. Given that TNFα induces FOXO3 nuclear exclusion, we hypothesized that FOXO3 directs TNF-induced transcriptional changes in FLS. Therefore, in line with a previous study [29] knockdown of FOXO3 likely reveals the contribution of this transcription factor to TNF-induced gene expression. RA-FLS were transfected with FOXO3 targeting siRNA pools. Then, FLS were stimulated with TNFα for 1 and 6 hours to dissect FOXO3 effects on early and late response genes (Fig. 3a). Within 1 hour, TNFα increased a number of well-known pro-inflammatory cytokines/chemokines (e.g. IL6, CCL20), proteolytic enzymes (e.g. MMP3) and inflammatory transcription factors (e.g. IRF1). A more profound up- and downregulation of genes was observed in FLS after 6 hours of TNFα stimulation (Fig. 3c). Consistent with the known function of TNFα to induce “interferon regulated genes” (IRGs) via de novo synthesis of IFNβ [30], the top genes displaying the highest degree of expression at 6 hours were the IRGs CXCL9, CXCL10, and CXCL11. These genes were not affected by short term (1 hour) treatment with TNFα and thus represent late response genes.
FOXO3-regulated gene expression in TNFα stimulated FLS. a Workflow outlining the RNA-sequencing (RNA-seq) experiment. RA-FLS from four different donors were transfected with non-targeting (siCTRL) or FOXO3-targeting siRNA pools and then stimulated with TNFα (10 ng/ml). RNA was isolated and processed for transcriptome profiling. b Representative western blots of RA-FLS after transfection with non-targeting (siCTRL) or FOXO3-targeting siRNA pools. c Volcano plots showing the magnitude of differential expressed genes. Bold numbers represent the amount of differential expressed genes (fold-change >2; Bonferroni corrected p-value <0.05; dispersion in replicates <0.2). d, e Scatter-plots showing the impact of FOXO3 knockdown on TNF-regulated genes. Dots with a text label represent TNF-regulated genes that are positively or negatively affected by FOXO3 silencing. Colors of the larger dots represent the temporal behavior of genes (for further definition see Supplementary Figure 1)
Silencing of FOXO3 mainly affected the expression of late TNFα response genes (Fig. 3d, e and Supplementary Fig. 1a). Most strikingly, FOXO3 silencing was mirrored by increased expression of 14 genes, including CXCL9, CXCL10, CXCL11 (which all bind to CXCR3), and TNFSF18 (GITRL). Gene ontology (GO) term analysis of these 14 genes revealed functions in leukocyte chemotaxis, especially for T-cells and monocytes (Supplementary Figure 1b). Importantly, we observed that siRNA-targeted reduction was more efficient to inactivate FOXO3 when compared to TNFα stimulation (Supplementary Fig. 2). Moreover, FOXO3 levels after siRNA mediated silencing were negatively correlated with the TNF-induced expression of CXCL10 (Supplementary Fig. 3). RNA-sequencing results for genes that were or were not affected by FOXO3 silencing were confirmed by qPCR and ELISA (Fig. 4a, b). Overall these data suggest that partial FOXO3 inactivation by TNFα due to nuclear exclusion is critical for the sustained expression of distinct pro-inflammatory chemokines in RA-FLS (Fig. 4a). By contrast, ELISA results did not support a role of FOXO3 in the regulation of IL6 and IL8 (Fig. 4b), suggesting that FOXO3 differentially regulates TNF-induced gene programs.
FOXO3 is a negative regulator of CXCR3-binding chemokines. a, b RA-FLS were transfected with non-targeting (siCTRL) or FOXO3-targeting siRNA pools. a Transfected FLS from six donors with RA were treated with TNFα (10 ng/ml) for 6 h. Gene expression was determined by qPCR and expression is presented relative to unstimulated cells. Values are the mean ± SEM. *p < 0.05, Wilcoxon matched pairs test. b Transfected RA-FLS (n = six) were treated with TNF (10 ng/ml) for 24 hours. Supernatants were analyzed by ELISA. Values are the mean ± SEM. *p < 0.05, Wilcoxon matched pairs test. c RA-FLS (n = 6) were pre-treated with DMSO, Ipatasertib (GDC-0068; 250 nM), Idelalisib (CAL-101; 250 nM), or Duvelisib (IPI-145; 250 nM) for 1 hour and stimulated with TNFα (10 ng/ml) for 24 hours. Release of CXCL10 was assessed by ELISA. Wilcoxon matched pairs test, *p < 0.05. d–f RA-FLS were pre-incubated with DMSO or Torin (250 nM) for 1 hour and stimulated with TNFα (10 ng/ml) for four (D. and E) or 24 hours (F.) d Representative western blots of at least four experiments with different RA-FLS cell lines. e Representative immunofluorescence images of FOXO3-localization (red) in RA-FLS. Nuclei were stained with DAPI (blue). Phalloidin was used to visualize actin (green). f CXCL10 release was measured by ELISA. Values are the mean ± SEM. *p < 0.05, Wilcoxon matched pairs test. n = 4
TNFα signaling co-opts the PI3K/AKT/mTOR/FOXO3 pathway to bring forth an IFN response
Nuclear exclusion (and thereby inactivation) of FOXO3 is induced by phosphorylation through the PI3K/AKT signaling pathway which is activated by TNFα [13, 27]. Therefore, inhibition of either, PI3K or AKT, would prevent TNF-induced FOXO3 phosphorylation and nuclear exclusion. Indeed, inhibition of PI3K-activity with the two highly specific inhibitors Idelalisib and Duvelisib prevented the TNF-induced inactivation of FOXO3 (Supplementary Fig. 4) and suppressed the TNF-induced expression of CXCL10 (Fig. 4c). Consistently, we found that the AKT-inhibitor Ipatasertib decreased FOXO3 phosphorylation (Supplementary Fig. 4) and CXCL10 production in TNF-stimulated fibroblasts (Fig. 4c). Previously we have shown that TNF-induced activation of AKT is dependent on mTOR [27]. To study whether mTOR is involved in the TNF-induced inactivation of FOXO3 we used the specific mTOR inhibitor Torin-1, which blocked the phosphorylation of the well-known mTOR substrate S6K (Fig. 4d). Torin-1 also prevented the phosphorylation (Fig. 4d) and nucleocytoplasmic shuttling of FOXO3 (Fig. 4e). Moreover, we observed decreased expression of CXCL10 (Fig. 4f), CXCL9 and TNFSF18 (data not shown) in Torin-1 treated RA-FLS. These data confirm the RNA-seq results and suggest that PI3K/AKT/mTOR act upstream of FOXO3 in TNF-stimulated FLS.
TNF-induced inactivation of phosphoinositide-3-kinase interacting protein 1 (PIK3IP1) promotes IRG expression in RA-FLS
Western blots and IF microscopy revealed a biphasic pattern of FOXO3 inactivation in TNF-stimulated FLS (Fig. 2a, c) with a strong, sustained second wave of FOXO3 phosphorylation and nuclear exclusion, which promotes the TNF-induced IFN-response. To unfold the mechanisms that contribute to the sustained inactivation of FOXO3 in response to TNF, we examined the RNA-seq data for TNF-driven regulators of the PI3K-AKT-FOXO3 pathway. Most strikingly, we found reduced expression of the PI3K-inhibitor PIK3IP1 [31] in TNF-treated FLS (Fig. 5a). Supporting the findings of others that PIK3IP1 is a direct target gene of FOXO3 [32], we observed decreased PIK3IP1 transcription in FOXO3 knockdown FLS (Fig. 5a, b). Time course experiments confirmed the RNA-seq data. PIK3IP1 expression was significantly reduced after 1 hour of TNFα stimulation and further decreased over time (Fig. 5c). Consistent with changes at the mRNA level, TNFα also caused downregulation of PIK3IP1 protein levels (Fig. 5d). To establish PIK3IP1 as a regulator of the TNF-induced IFN-response we used siRNA pools to silence its expression in FLS. Knockdown of PIK3IP1 was confirmed by qPCR (data not shown) and western blotting (Fig. 5e). Of note, targeted reduction by siRNA was more efficient to reduce PIK3IP expression than TNFα stimulation (Supplementary Fig. 5a). As expected, PIK3IP1 silencing reduced the nuclear levels of FOXO3 (Fig. 5f). Most strikingly, TNF-induced expression of CXCL9, CXCL10, CXCL11, and TNFSF18 was increased in FLS that were transfected with PIK3IP-targeting siRNA pools (Figs. 5g, h). Moreover, we observed an inverse correlation between PIK3IP cellular levels and TNF-induced IRG expression (Supplementary Fig. 5b). Taken together, these data suggest that TNF-mediated inactivation of FOXO3 limits the transcription of PIK3IP1, which leads to sustained activation of PI3K/AKT signaling and enhanced expression of CXCL9, CXCL10, CXCL11, and TNFSF18 (Fig. 6).
An important role for the FOXO3 target PIK3IP in TNF-induced fibroblast-activation. a Heatmap of PI3K/AKT pathway regulating genes that are responsive to TNFα stimulation and siFOXO3 mediated knockdown. Genes are clustered by complete hierarchical clustering with 1 - Pearson correlation as distance. Green labeled genes are positive, whereas red labeled genes are negative regulators of the PI3K/AKT pathway. b FLS were transfected with non- (siCTRL) or FOXO3-targeting siRNA pools. Gene expression was determined by qPCR and expression is presented relative to GAPDH. Values are the mean ± SEM. *p < 0.05, Wilcoxon matched pairs test. n = 6. c PIK3IP expression in TNF-treated (10 ng/ml) RA-FLS (n = 4). Gene expression was determined by qPCR and expression is presented relative to untreated cells. d Western blots of PIK3IP expression in TNFα (10 ng/ml) stimulated FLS. Representative blots of four independent experiments. e–h FLS were transfected with non- (siCTRL) or PIK3IP1-targeting siRNA pools. e Representative western blots of PIK3IP1 expression in transfected FLS. f Representative immunofluorescence images of FOXO3 in siRNA transfected FLS. g Transfected RA-FLS (n = 5) were stimulated with TNFα (10 ng/ml) for 6 hours. Gene expression was determined by qPCR. Expression is presented relative to unstimulated cells. Values are the mean ± SEM. *p < 0.05, **p < 0.01. Wilcoxon matched pairs test. h CXCL10 expression of TNFα stimulated FLS (10 ng/ml, 24 h). *p < 0.05, Wilcoxon matched pairs test. n = 6
A model depicting the TNF/PI3K/AKT/FOXO3/PIK3IP1 signaling circuits in RA-FLS. In FLS TNF activates the PI3K/AKT pathway leading to inactivation of FOXO3. FOXO3 inactivation promotes the TNF-induced expression of IRGs, such as CXCL9, CXCL10, CXCL11, and TNFSF18. TNF stimulation also decreases the transcription of PIK3IP1, which further promotes PI3K/AKT activation and FOXO3 inactivation, thereby amplifying the expression of IRGs
Discussion
Perturbations in the expression or activity of FOXO3 are increasingly linked to chronic inflammatory diseases, such as colitis and arthritis [14, 15, 33, 34]. Research into the mechanism of the FOXO3-regulated inflammatory response has mainly concentrated on leukocytes. Thus, critical roles for FOXO3 were shown for T-cell proliferation and activation [35], monocyte cytokine secretion [15], and neutrophil survival [36]. Here we show that FOXO3 also directs the inflammatory response in non-immune cells, namely FLS. This is particularly important since current models increasingly stress the role of mesenchymal stromal cells in non-resolving inflammatory diseases, such as RA [37].
In accordance with previous studies [10, 34] we found phosphorylation/inactivation of FOXO3 in the inflamed, rheumatoid synovium. Intriguingly, there was no difference in total FOXO3 expression between RA and OA synovial tissues, suggesting that the inflammatory response is primarily regulated by FOXO3 phosphorylation, but not by altered gene expression. While these preceding studies highlighted the strong phosphorylation of FOXO3 in leukocytes, we noted that FOXO3 is primarily phosphorylated in fibroblast-like cells of the synovial membrane. Based on this observation we hypothesized that FOXO3 is inactive in TNF-stimulated FLS. Indeed, TNFα, which is one of the main drivers of FLS activation in RA [3], induced partial FOXO3 phosphorylation and nuclear exclusion in these cells. Given the strong phenotype in the IHC staining, other pro-inflammatory mediators may also induce FOXO3 inactivation in the inflamed synovium. To address the FOXO3 contribution to the inflammatory activity of FLS we performed gain-of-function (pharmacological inhibition of the FOXO3 upstream kinases AKT, mTOR, and PI3K) as well as loss-of-function (siRNA mediated knockdown of FOXO3) experiments. Strikingly, we found that FOXO3 activity is mirrored by the expression of a subset of genes involved in the TNF-induced interferon response, such as CXCL9, CXCL10, CXCL11, and TNFSF18. Numerous studies have shown that these inflammatory mediators are increased in RA synovial tissues and fluids [3, 38,39,40,41]. CXCL10, as well as the other CXCR3-binding chemokines, play a major role in leukocyte homing to inflamed tissues and thereby contribute to the perpetuation of inflammation [42]. TNFSF18 is the ligand for TNFRSF18/AITR/GITR and is best known to regulate T-cell responses by lowering the threshold for T-cell activation and proliferation [43]. Thus, our data suggest that TNF-induced FOXO3 inactivation in FLS particularly promotes leukocyte recruitment to the synovium and local T-cell activation. By contrast, we observed decreased expression of MMP3 and CSF2 in FOXO3 knockdown FLS. Thus, signaling events upstream of FOXO3, such as PI3K and AKT activation, resulting in FOXO3 nuclear exclusion might have beneficial (e.g. inhibition of CXCL10 expression) but also detrimental effects by increasing the secretion of MMP3 and CSF2. This has to be considered in future clinical trials investigating inhibition of the PI3K/AKT pathway.
But how does FOXO3 regulate the TNF-induced IFN-response in FLS? The promoters of TNF-induced IFN-response genes are not enriched for potential FOXO3 binding sites as suggested by bioinformatics analysis (data not shown). Thus, FOXO3 does not directly regulate these genes. In macrophages FOXO3 was shown to suppress the antiviral response by directly affecting the IRF7-dependent expression of IFNβ [29]. A similar mechanism may operate in TNF-stimulated FLS. TNF-stimulated FLS secrete low amounts of IFNβ, which acts in an autocrine/paracrine manner to activate the transcription factor STAT1, which induces IRG expression [30]. We indeed observed higher STAT1 activity in FOXO3-knockdown FLS (Supplementary Fig. 6). Correspondingly the RNA-seq results showed increased expression of IRF7 and IFNβ in FOXO3 knockdown FLS (data not shown). However, the difference was statistically not significant and further experiments are needed to proof these observations.
More importantly, we herein identify PIK3IP1 as a yet unknown regulator of the cellular response to TNFα. Our experimental data demonstrate that TNFα induces FOXO3-inactivation, which represses the expression of the FOXO3 transcriptional target PI3KIP1. Reduced PI3KIP3 results in increased PI3K/AKT pathway activity, which supports the expression of CXCL9, CXCXL10, CXCL11, and TNFSF18 in FLS. PIK3IP1 is a tumor suppressive protein [44, 45] that functions as a direct inhibitor of the PI3K [31]. PIK3IP has also been implicated in T-cell activation [46, 47].
Overall, our study identifies a previously unknown TNF-activated signaling circuit that contributes to the TNF-induced IFN-response. Thus, our work on FOXO3 and its transcriptional target PIK3IP1 as important modulators for FLS-mediated inflammation provides novel insights into the molecular mechanism underlying TNF-driven inflammatory diseases.
References
Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388:2023–2038.
Noss EH, Brenner MB. The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunol Rev. 2008;223:252–70.
Jones DS, Jenney AP, Swantek JL, Burke JM, Lauffenburger DA, Sorger PK. Profiling drugs for rheumatoid arthritis that inhibit synovial fibroblast activation. Nat Chem Biol. 2017;13:38–45.
Bartok B, Boyle DL, Liu Y, Ren P, Ball ST, Bugbee WD, et al. PI3 kinase delta is a key regulator of synoviocyte function in rheumatoid arthritis. Am J Pathol. 2012;180:1906–16.
Hayer S, Pundt N, Peters MA, Wunrau C, Kühnel I, Neugebauer K, et al. PI3Kgamma regulates cartilage damage in chronic inflammatory arthritis. FASEB J. 2009;23:4288–98.
Zhang HG, Wang Y, Xie JF, Liang X, Liu D, Yang P, et al. Regulation of tumor necrosis factor alpha-mediated apoptosis of rheumatoid arthritis synovial fibroblasts by the protein kinase Akt. Arthritis Rheum. 2001;44:1555–67.
Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–35.
Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405.
Cejka D, Hayer S, Niederreiter B, Sieghart W, Fuereder T, Zwerina J, et al. Mammalian target of rapamycin signaling is crucial for joint destruction in experimental arthritis and is activated in osteoclasts from patients with rheumatoid arthritis. Arthritis Rheum. 2010;62:2294–302.
Ludikhuize J, de Launay D, Groot D, Smeets TJ, Vinkenoog M, Sanders ME, et al. Inhibition of forkhead box class O family member transcription factors in rheumatoid synovial tissue. Arthritis Rheum. 2007;56:2180–91.
Camps M, Rückle T, Ji H, Ardissone V, Rintelen F, Shaw J, et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–43.
Randis TM, Puri KD, Zhou H, Diacovo TG. Role of PI3Kdelta and PI3Kgamma in inflammatory arthritis and tissue localization of neutrophils. Eur J Immunol. 2008;38:1215–24.
Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14:83–97.
Viatte S, Lee JC, Fu B, Espéli M, Lunt M, De Wolf JN, et al. Association between genetic variation in FOXO3 and reductions in inflammation and disease activity in inflammatory polyarthritis. Arthritis Rheum. 2016;68:2629–36.
Lee JC, Espeli M, Anderson CA, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69.
Johnson DM, Hayat SQ, Burton GV. Rheumatoid arthritis complicating adjuvant interferon-alpha therapy for malignant melanoma. J Rheumatol. 1999;26:1009–10.
Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–81.
Kiener HP, Lee DM, Agarwal SK, Brenner MB. Cadherin-11 induces rheumatoid arthritis fibroblast-like synoviocytes to form lining layers in vitro. Am J Pathol. 2006;168:1486–99.
Rosner M, Siegel N, Fuchs C, Slabina N, Dolznig H, Hengstschläger M. Efficient siRNA-mediated prolonged gene silencing in human amniotic fluid stem cells. Nat Protoc. 2010;5:1081–95.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–9.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29.
Karonitsch T, Kandasamy RK, Kartnig F, Herdy B, Dalwigk K, Niederreiter B, et al. mTOR senses environmental cues to shape the fibroblast-like synoviocyte response to inflammation. Cell Rep. 2018;23:2157–67.
Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91.
Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, et al. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2012;490:421–5.
Rosengren S, Corr M, Firestein GS, Boyle DL. The JAK inhibitor CP-690,550 (tofacitinib) inhibits TNF-induced chemokine expression in fibroblast-like synoviocytes: autocrine role of type I interferon. Ann Rheum Dis. 2012;71:440–7.
Zhu Z, He X, Johnson C, Stoops J, Eaker AE, Stoffer DS, et al. PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein. Biochem Biophys Res Commun. 2007;358:66–72.
Schmidt-Strassburger U, Schips TG, Maier HJ, Kloiber K, Mannella F, Braunstein KE, et al. Expression of constitutively active FoxO3 in murine forebrain leads to a loss of neural progenitors. FASEB J. 2012;26:4990–5001.
Snoeks L, Weber CR, Wasland K, Turner JR, Vainder C, Qi W, et al. Tumor suppressor FOXO3 participates in the regulation of intestinal inflammation. Lab Invest. 2009;89:1053–62.
Turrel-Davin F, Tournadre A, Pachot A, Arnaud B, Cazalis MA, Mougin B, et al. FoxO3a involved in neutrophil and T cell survival is overexpressed in rheumatoid blood and synovial tissue. Ann Rheum Dis. 2010;69:755–60.
Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–13.
Jonsson H, Allen P, Peng SL. Inflammatory arthritis requires Foxo3a to prevent Fas ligand-induced neutrophil apoptosis. Nat Med. 2005;11:666–71.
Buckley CD. Why does chronic inflammation persist: aan unexpected role for fibroblasts. Immunol Lett. 2011;138:12–14.
Patel DD, Zachariah JP, Whichard LP. CXCR3 and CCR5 ligands in rheumatoid arthritis synovium. Clin Immunol. 2001;98:39–45.
Bae E, Kim WJ, Kang YM, Suk K, Koh EM, Cha HS, et al. Glucocorticoid-induced tumour necrosis factor receptor-related protein-mediated macrophage stimulation may induce cellular adhesion and cytokine expression in rheumatoid arthritis. Clin Exp Immunol. 2007;148:410–8.
Ueno A, Yamamura M, Iwahashi M, Okamoto A, Aita T, Ogawa N, et al. The production of CXCR3-agonistic chemokines by synovial fibroblasts from patients with rheumatoid arthritis. Rheumatol Int. 2005;25:361–7.
Yoshida S, Arakawa F, Higuchi F, Ishibashi Y, Goto M, Sugita Y, et al. Gene expression analysis of rheumatoid arthritis synovial lining regions by cDNA microarray combined with laser microdissection: up-regulation of inflammation-associated STAT1, IRF1, CXCL9, CXCL10, and CCL5. Scand J Rheumatol. 2012;41:170–9.
Lee EY, Lee ZH, Song YW. The interaction between CXCL10 and cytokines in chronic inflammatory arthritis. Autoimmun Rev. 2013;12:554–7.
Tone M, Tone Y, Adams E, Yates SF, Frewin MR, Cobbold SP, et al. Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells. Proc Natl Acad Sci USA. 2003;100:15059–64.
He X, Zhu Z, Johnson C, Stoops J, Eaker AE, Bowen W, et al. PIK3IP1, a negative regulator of PI3K, suppresses the development of hepatocellular carcinoma. Cancer Res. 2008;68:5591–8.
Wong CC, Martincorena I, Rust AG, Rashid M, Alifrangis C, Alexandrov LB, et al. Inactivating CUX1 mutations promote tumorigenesis. Nat Genet. 2014;46:33–38.
Wei H, Geng J, Shi B, Liu Z, Wang YH, Stevens AC, et al. Cutting edge: Foxp1 controls naive CD8+T cell quiescence by simultaneously repressing key pathways in cellular metabolism and cell cycle progression. J Immunol. 2016;196:3537–41.
DeFrances MC, Debelius DR, Cheng J, Kane LP. Inhibition of T-cell activation by PIK3IP1. Eur J Immunol. 2012;42:2754–9.
Acknowledgements
This work was supported by the Innovative Medicines Initiative Joint Undertaking under grant agreement #115142 (BTCure) and #777357 (RTCure), the “Rheumapreis des St. Anna Kinderspitals“ and the Vienna Science and Technology Fund (WWTF #LS13-092).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Brandstetter, B., Dalwigk, K., Platzer, A. et al. FOXO3 is involved in the tumor necrosis factor-driven inflammatory response in fibroblast-like synoviocytes. Lab Invest 99, 648–658 (2019). https://doi.org/10.1038/s41374-018-0184-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0184-7
This article is cited by
-
Screening crucial lncRNAs and genes in osteoarthritis by integrated analysis
Advances in Rheumatology (2023)
-
Down-Regulation of microRNA-30d Alleviates Intervertebral Disc Degeneration Through the Promotion of FOXO3 and Suppression of CXCL10
Calcified Tissue International (2021)