Abstract
The roles and downstream target genes of the transcription factor ZNF326 in malignant tumors are unclear. Out of 146 lung cancer tissue samples, we found that high expression of ZNF326 in 82 samples was closely related to low differentiation and a high pTNM stage of non-small cell lung cancer (NSCLC) cells. In vitro and in vivo analyses showed that ZNF326 significantly promoted cell cycle progression, colony formation, and proliferation as well as the growth of NSCLC transplanted tumors. Chromatin immunoprecipitation sequencing, dual-luciferase assay, and electrophoretic mobility shift assay confirmed that the C2H2 structure of ZNF326 binds to the −833 to −875 bp region of the ERCC1 promoter to initiate transcriptional activity. This binding promoted CyclinB1 synthesis and cell cycle progression. These results show that the ZNF326 transcription factor is highly expressed in lung cancer and promotes the proliferation of NSCLC cells by regulating the expression of ERCC1.
Similar content being viewed by others
Introduction
Lung cancer has high morbidity and mortality worldwide [1,2,3]. Recent studies have shown that many zinc finger proteins are closely related to the occurrence and progression of lung cancer. For example, ZNF322A transcriptionally deregulates alpha-adducin, CyclinD1, and p53 to promote tumor growth and metastasis in lung cancer [4]. The zinc finger transcription factor ZFX promotes the growth and migration of non-small cell lung cancer (NSCLC) cells [5]. ZNF326 is a newly discovered C2H2 zinc finger protein. Studies on ZNF326 are limited. Rangel et al. [6] reported that ZNF326 exhibits low expression in triple-negative breast cancer cell lines, and ZNF326 knockdown not only upregulates the expression of Epithelial-mesenchymal transition (EMT)- and tumor stem cell-associated genes but also promotes tumorigenesis in nude mice. Rengasamy et al. [7] found that PRMT5/WDR77 forms a complex with methylated ZNF326 in breast cancer to regulate splicing and maintain stability of genes such as REPIN1, AP4, ST3GAL6, TRNAU1AP, and PFKM, involved in breast cancer cell proliferation and migration. Close et al. reported that deleted in breast cancer-1 and ZNF326 form complexes called DBIRD. DBIRD binds directly to RNAPII and regulates the alternative splicing of a large set of exons embedded in (A + T)-rich DNA [8]. It is well known that different zinc finger proteins play various roles in promoting or inhibiting cancer and that a particular zinc finger protein may have opposite roles in different tumors. However, in-depth analyses of whether ZNF326 promotes or suppresses cancer and the mechanisms underlying its effects are lacking.
Znf326 is located at 1p22. 2 and is 404,376 kb in length. The Znf326 protein is composed of 582 amino acids of approximately 75 kDa and includes an N-terminal transcriptional activation region (59–124 aa), nuclear locus signal (237–261 aa), two C2H2 zinc finger structures (314–336 aa and 407–430 aa), and a C-terminal glutamate-rich sequence [9]. These structural features enable Znf326 to act as a transcription factor; however, its target genes and regulatory mechanisms are unknown.
In this study, chromatin immunoprecipitation sequencing (ChIP-seq) was used to sequence and analyze DNA fragments associated with the ZNF326 antibody and Myc-tag antibody, and potential target genes in the promoter region of the DNA fragment were screened. After alteration in the expression of ZNF326, the mRNA levels of potential target genes were measured by quantitative reverse transcription polymerase chain reaction (RT-qPCR). Electrophoretic mobility shift assay (EMSA) and dual-luciferase assays were used to identify the specific binding sequences. The results of in vitro and in vivo analyses revealed the molecular mechanism by which ZNF326 functions as a transcription factor.
Materials and methods
Patient sample information and cell culture conditions
In total, 146 NSCLC specimens (in addition to 60 matched normal lung tissues) were collected from the Department of Pathology of the First Affiliated Hospital of China Medical University. No patient received chemotherapy or radiotherapy before surgery. Based on 2015 World Health Organization classification guidelines [10], there were 84 cases of adenocarcinoma and 62 cases of squamous cell carcinoma. Lung tumors were differentiated based on the following criteria: lepidic adenocarcinoma is well-differentiated adenocarcinoma; acinar or papillary adenocarcinoma is considered moderately differentiated adenocarcinoma; solid and micropapillary adenocarcinoma is classified as poorly differentiated adenocarcinoma; well-differentiated squamous cell carcinoma shows obvious keratinization, pearl formation, or intercellular bridges; the cell nuclei typically have dark, non-transparent chromatin, without prominent nucleoli; keratinization is present only focally or less apparent in moderately differentiated squamous cell carcinoma; cytoplasmic keratinization in poorly differentiated squamous cell carcinoma is absent or inapparent, and nuclei may have open chromatin with prominent nucleoli. According to the International Anti-Cancer Union (TNM) staging criteria of 2017 [11], there were 125 cases of stage I–II and 21 cases of stage III. This study was performed with the approval of the Ethics Committee of the China Medical University (No. 2015[LS]023, China Medical University).
A549 and H1299 lung cancer cell lines were purchased from the Shanghai Cell Bank. The cells were cultured in accordance with the culture conditions recommended by the Shanghai Cell Bank. All cell lines were authenticated by short tandem repeat DNA profiling.
Immunoblotting analysis
Total protein (60 µg) was extracted using radioimmunoprecipitation assay pyrolysis solution. Proteins were separated by 8–10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% bovine serum albumin for 2 h at room temperature and incubated with antibodies overnight at 4 °C. After washing, the membranes were incubated with peroxidase-conjugated anti-mouse/rabbit IgG at 37 °C for 2 h. The relative protein levels were calculated using GAPDH as a loading control. The bands were quantified using ImageJ software. Antibody details are listed in Supplementary Table S1.
Plasmids, short hairpin RNA, small interfering RNA, and transfection
G418 (#A1720; Sigma, St. Louis, MO, USA) was used to screen stably transfected cells. Information on plasmids, small interfering RNA, short hairpin RNA (shRNA), and ZNF326 constructs is provided in Supplementary Table S2. Plasmids and shRNA were delivered using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Immunohistochemistry
Paraffin-embedded specimens were cut into sections with a thickness of 4 μm. The sections were stained by the S-P immunohistochemical method. Tissues were incubated with antibodies (ZNF326, HPA028466, 1:200, Sigma; ERCC1, 8F1, 1:100, Abcam, Cambridge, UK). The intensity of ZNF326 and ERCC1 staining was scored as follows: 0 (no staining), 1 (weak), 2 (moderate), and 3 (high). Percentage scores were assigned as follows: 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The scores for each tumor sample were multiplied to obtain a final score of 0–12. Tumor samples with scores of ≥4 were considered to show high ZNF326 expression, and tumor samples with scores between 0 and 4 were considered to show low ZNF326 expression. Phosphate-buffered saline and goat serum were used as negative controls (MaiXin, Fuzhou, China).
Dual-luciferase assay
p-ERCC1-GL3 (−1800 bp), p-ERCC1-GL3-ΔR1 (−859 to −851 + −833 to −825 bp deleted), p-ERCC1-GL3--ΔR2 (−883 to −875 bp deleted), p-ERCC1-GL3--ΔR1&ΔR2 (−859 to −851 + −833 to −825 + −883 to −875 bp deleted), and p-RL-TK Luciferase reporter genes were synthesized by the Shanghai Jima Company. The Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA) was used for detection.
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium experiment, colony formation, and Matrigel invasion assays
Cells (3000 cells/well) were seeded in a 96-well plate. Cell proliferation was detected using 3-(4,5-dimethylthiazol-2-yl)−5-(3-carboxymethoxyphenyl)−2-(4-sulfophenyl)−2H-tetrazolium (MTS; Promega). MTS was added to each well at 1:10 (v/v) per 100 µl and incubated for 2 h at 37 °C. The results were quantitated by spectrophotometry using a test wavelength of 490 nm.
A total of 500 cells were seeded in a 6-cm culture dish for 14 days. Cells were then fixed with methanol and stained with hematoxylin. Colonies with more than 50 cells were counted.
Cells (5 × 105) were seeded into the upper Transwell® chambers with a pore size of 8 μm (Costar, Cambridge, MA, USA) with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) and cultured for 24 h. Cells were then fixed with methanol and stained with hematoxylin. Ten fields were randomly selected, and the number of cells invading the chamber was counted.
RT-qPCR assay
The ReliaPrep RNA Cell MiniPrep system was used to extract total RNA from cells, and reverse transcription was performed using the GoScript Reverse Transcription system with 20 µl. qPCR Master Mixer and ABI7900 Real-Time Quantification were used under the following conditions: 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 60 s for 40 cycles (Promega). The relative transcript levels of genes were normalized to GAPDH mRNA levels (primer sequences are shown in Supplementary Table S3).
Flow cytometric assay
Cells were digested using trypsin. The cell concentration was adjusted to 2 × 106 with phosphate-buffered saline. Samples were supplemented with 70% cold ethanol, fixed overnight, treated with RNaseA at 37 °C in a water bath for 30 min, stained with propidium iodide in the dark for 30 min at 4 °C, and analyzed by flow cytometry using the FACSCalibur flow cytometer with the CellQuest 3.0 software (BD Biosciences, Billerica, MA, USA).
Transplantation of tumor cells into nude mice
The nude mice used in this study were treated according to the experimental animal ethics guidelines issued by the China Medical University. The study was approved by the Institutional Animal Research Committee of China Medical University. Nude mice (4–6 weeks old) were purchased from Beijing Weitonglihua Company (Beijing, China). ZNF326-overexpressing or -silenced cells in each group were adjusted to 5 × 106/ml, and 0.2 ml of the cell solution was injected subcutaneously into nude mice. The mice were sacrificed after 6 weeks, and the tumors were excised and weighed.
Chromatin co-precipitation and sequencing
ZNF326-overexpressing H1299 cells were crosslinked and lysed by ultrasound treatment. The pyrolysis liquid was divided into four groups and treated with different antibodies. Protein G agarose was added at 4 °C, followed by rotation for 1 h. The corresponding antibody was then added, followed by incubation overnight at 4 °C. After protein/DNA complex elution, the complex was disintegrated. After recovery of DNA samples, high-throughput sequencing was performed (17–371; Millipore, Burlington, MA, USA).
Electrophoretic mobility shift assay
Nuclear protein was extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher, Waltham, MA, USA), and 4–8 µg of cell nuclear protein was used for the EMSA system binding reaction. The prepared mixture was electrophoresed through 6% non-denatured gel and subsequently transferred to a membrane. After crosslinking with a 254-nm ultraviolet lamp tube, samples were blocked and washed. Finally, the biotin-labeled probe was detected by luminescence (Thermo Fisher). The probes are listed in Supplementary Table S4.
Statistical analysis
All data were analyzed using the SPSS statistical software version 22.0 (SPSS, Chicago, IL, USA). The correlation between ZNF326 expression and clinicopathological factors was examined by Chi-squared tests, and that between ZNF326 and ERCC1 levels was analyzed based on Spearman correlation coefficients. Two-tailed Student’s t-tests were used to evaluate the results of cell and animal studies. Each experiment was repeated at least three times. p < 0.05 was considered statistically significant.
Results
ZNF326 promotes proliferation of NSCLC cells in vivo and in vitro
To explore the biological function of ZNF326 in NSCLC, we used H1299 cell lines with stable ZNF326 expression and with ZNF326 knockdown (selected by G418) to evaluate tumorigenesis in nude mice. The volume and weight of transplanted tumors in the injection group with cells overexpressing ZNF326 were significantly greater than those in the control group (95 ± 10.000 vs. 246.667 ± 11.015 mm3, p < 0.01; 0.206 ± 0.01 vs. 0.442 ± 0.06 g, p < 0.05; Fig. 1a). ZNF326 knockdown had the opposite effect (322 ± 14.107 vs. 253 ± 21.517 mm3, p < 0.05; 0.426 ± 0.009 vs. 0.227 ± 0.007 g, p < 0.01; Fig. 1b). Next, we verified the effect of the proliferation of NSCLC cells by bidrectional regulation of ZNF326 expression in vitro. In H1299 and A549 cells, ZNF326 overexpression promoted the cell proliferation as evidenced by the MTS assay and colony formation. Consistent with these observations, ZNF326 knockdown inhibited cell proliferation and colony formation (Fig. 1c–e).
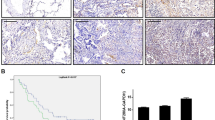
ZNF326 promotes proliferation of non-small cell lung cancer (NSCLC) cells in vivo and in vitro. ZNF326 promotes proliferation in vivo. a Subcutaneous injection of H1299 cells stably expressing ZNF326 into nude mice; the volumes and weights of the transplanted tumors were greater in the injection group with cells overexpressing ZNF326 than that in the control group. b The volumes and weights of transplanted tumors containing the ZNF326 knockdown H1299 cells were lower than those in the control group. ZNF326 promotes proliferation in vitro. Effect of ZNF326 expression on proliferation and colony formation of NSCLC cells. H1299 cells and A549 cells were transfected with ZNF326 plasmid or control, or with ZNF326-specific shRNA or control shRNA. *p < 0.05, **p < 0.01. c, d MTS assay. H1299: NC vs. ZNF326, p = 0.001; shNC vs shZNF326, p = 0.003; A549: NC vs. ZNF326, p = 0.006; shNC vs shZNF326, p = 0.037. e Colony formation assays. H1299: NC vs. ZNF326, p = 0.000; shNC vs shZNF326, p = 0.000; A549: NC vs. ZNF326, p = 0.000; shNC vs shZNF326, p = 0.001. f The expression of ZNF326 was low in the bronchial epithelium and alveolar tissue (a) but high in the nuclei of adenocarcinoma cells (b) and in squamous cell carcinoma cells (c)
To confirm the results of in vitro experiments, we analyzed the expression of the ZNF326 protein in the 146 NSCLC tissues. We observed nuclear overexpression of ZNF326 in NSCLC tissues (82/146 and 6/60 paracancerous lung tissues, p < 0.05). The high expression of ZNF326 was closely related to low differentiation and a high pTNM stage of NSCLC. There was no significant correlation between ZNF326 expression and age, sex, and lymph node metastasis (Fig. 1f, Table 1). These results suggested that ZNF326 serves as a carcinogenic factor in the development of NSCLC.
ERCC1 is a target gene of ZNF326
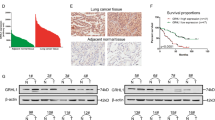
To address the molecular mechanism by which ZNF326 promotes the proliferation of cells in NSCLC, we used ChIP-seq to analyze ZNF326- and Myc-tag-binding DNA fragment. In total, 3848 DNA fragments were screened using a ZNF326 antibody, and 2558 DNA fragments were screened using a Myc-tag antibody. These DNA fragments were mainly located in promoter, exon, intron, and intergenic regions. We found 278 common DNA fragments (Fig. 2a). The broad peak pattern was used to analyze the protein-binding sequences. We screened 8 potential target genes (Table 2, Supplementary Table S5-S6). We subsequently assessed the mRNA expression of these potential target genes by RT-PCR in H1299 and A549 cells transfected with the ZNF326 plasmid. ZNF326 overexpression upregulated the mRNA level of ERCC1 and HDAC7 but downregulated that of ZNF383 and LTBP4. The mRNA level of CAPN12, ARHGAP33, CYP2A7, and DUX4L19 did not significantly change (Fig. 2b), whereas knockdown of ZNF326 had the opposite effects (Fig. 2c). Among the 8 potential target genes, the change in ERCC1 was the most obvious and stable. To further confirm that ERCC1 is a target gene of ZNF326, we assessed the effect of ZNF326 on the activities of ERCC1 using luciferase reporter assays. In H1299 and A549 cells, ZNF326 overexpression enhanced the activity of ERCC1. However, ZNF326 silencing in H1299 and A549 cells had the opposite effect (Fig. 2d).
ERCC1 is a target gene of ZNF326. a DNA fragments bound with ZNF326 antibody and Myc-tag antibody were mainly located in the promoter, exon, intron, and intergenic regions. A total of 278 common DNA fragments were screened. b Transfection of ZNF326 into H1299 and A549 cells upregulated the mRNA level of ERCC1, blocked ZNF326 protein expression, and downregulated the mRNA level of ERCC1. c, d ZNF326 overexpression enhanced the activity of ERCC1. However, ZNF326 silencing in H1299 cells and A549 cells had the opposite effect. Triplicate samples were analyzed for each treatment, and the results are presented as the mean ± SD, *p < 0.05, **p < 0.01. e ZNF326 was positively correlated with ERCC1 expression in NSCLC, which was consistent with the website prediction. LUAD lung adenocarcinoma, LUSC lung squamous carcinoma
In addition, ERCC1 expression was assessed by immunohistochemistry in 146 NSCLC samples. Compared with the ZNF326 immunohistochemistry staining results, the expression of ERCC1 was positively correlated with the expression of ZNF326 in NSCLC. These results are consistent with the prediction results of the http://gepia.cancer-pku.cn/ website (r = 0.737, p < 0.01; Fig. 2e, Table 3). These results showed that ERCC1 is a target gene of ZNF326 in NSCLC.
ZNF326 regulates the expression of ERCC1 via the −883 to −875 bp sequence in the ERCC1 promoter
We also examined how ZNF326 regulates ERCC1 expression. According to ChIP-seq and related studies [12], there are four sequences in the upstream region of ERCC1 that may interact with ZNF326, i.e., region 1 (−859 to −851 + −833 to −825 bp), region 2 (−883 to −875 bp), region 3 (−910 to −901 bp), and region 4 (−966 to −960 + −958 to −951 bp). We performed EMSA and observed that region 2 (−883 to −875 bp) combined with ZNF326 exhibited a specific migration band (Fig. 3a), while region 1 (−859 to −851 + −833 to −825 bp) exhibited a nonspecific migration band (Fig. 3b). Region 3 and region 4 exhibited no band. To further verify that region 2 (−883 to −875 bp) is the binding site of ZNF326, we constructed a series of ERCC1 promoter luciferase reporter gene mutant plasmids (Fig. 3c). Luciferase reporter assays showed that ZNF326 did not enhance the activity of the ERCC1 luciferase reporter gene after knockout of region 2 (Fig. 3d, e). In addition, according to previous studies [9, 12, 13], we designed a series of ZNF326 mutant plasmids (Fig. 3f). Luciferase reporter assays indicated that transcriptional activation domain (TAD) and Zn (C2H2 domain) were necessary for ZNF326 to activate ERCC1 transcription (Fig. 3g, h). These results demonstrated that Zn (C2H2 domain) of ZNF326 can combine with region 2 (−883 to −875 bp) of the ERCC1 promoter to promote ERCC1 transcription.
ZNF326 initiates the transcriptional activity of ERCC1 via its C2H2 structure, which binds to the region (−883 to −875 bp) of the ERCC1 promoter. a EMSA revealed that region 2 (−883 to −875 bp), combined with ZNF326, exhibited a specific migration band. b EMSA revealed that region 1 (−859 to −851 + −833 to −825 bp), combined with ZNF326, exhibited a nonspecific migration band. c Mutant plasmids of three ERCC1 promoter luciferase reporter genes. TSS transcription starting site. d, e The transcriptional activity of ERCC1 was only activated by preserving region 2 (−883 to −875 bp) of ZNF326. f Five ZNF326 mutant plasmids. TAD transcriptional activation domain, NLS nuclear locus signal, Zn1 the first C2H2 zinc finger structure, Zn2 the second C2H2 zinc finger structure, Glu-rich C-terminal glutamate-rich domain. g Luciferase reporter gene experiments showed that the TAD and Zn domain were necessary for ZNF326 to activate ERCC1 transcription. Triplicate samples were analyzed for each treatment, and the results are presented as the mean ± SD, *p < 0.05, **p < 0.01
ZNF326 regulates proliferation-related protein expression to affect the cell cycle
To elucidate the mechanism by which ZNF326 promotes the proliferation of NSCLC cells, we overexpressed ZNF326 in H1299 and A549 cells to assess the cell cycle by flow cytometry. The results are shown in Fig. 4a–d. The number of cells entering the G2/M phase increased after ZNF326 overexpression, but decreased after ZNF326 knockdown. To further explain this phenomenon, we measured changes in the expression of key proteins as the G2/M stage of the cell cycle progressed, and we found that the expression of CyclinA2 and CyclinB1 was upregulated and that of P21 was downregulated by ZNF326 overexpression in A549 and H1299 cells, whereas knockdown of ZNF326 had the opposite effects (Fig. 4e, f).
ZNF326 regulates proliferation-related protein expression to affect the cell cycle. a–d Flow cytometric analysis. Overexpression of ZNF326 increased the number of cells transitioning from S phase to G2/M phase, which was decreased after knockdown of ZNF326. Triplicate samples were analyzed for each treatment, and the results are presented as the mean ± SD, *p < 0.05, **p < 0.01. e, f H1299 cells and A549 cells were transfected with ZNF326 plasmid or control, or with ZNF326-specific shRNA or control shRNA. Changes in expression of cell proliferation-related proteins in H1299 (e) cells and A549 (f) cells
ZNF326 promotes CyclinB1 protein synthesis via ERCC1
It was not clear whether the promotion of proliferation was related to ERCC1 or due to the direct effect of ZNF326. Therefore, we confirmed the relationship by transfection with ZNF326-ΔTAD or ZNF326-ΔZn, which affected the expression of ERCC1. Immunoblotting showed that after transfection with ZNF326-ΔTAD or ZNF326-ΔZn mutant plasmids, the level of ERCC1 decreased significantly, and that of CyclinB1 also decreased. Moreover, the changes in CyclinA2 and P21 were not significantly related to the changes in ERCC1 protein levels (Fig. 5a). These results suggested that ZNF326 may directly regulate CylinB1 via ERCC1. However, the regulation of CyclinA2 and P21 cannot be achieved through ERCC1. Co-transfection of the ZNF326 and ERCC1 plasmids indicated that ZNF326 and ERCC1 had a synergistic effect on the upregulation of CyclinB1 (Fig. 5b). Interfering with ERCC1 in addition to ZNF326 plasmid transfection abolished the synergistic effect (Fig. 5c). The colony formation assays also confirmed that ERCC1 and ZNF326 both promoted H1299 and A549 cell colony formation (Fig. 5d–g). These results suggested that ZNF326 positively regulates the expression of CyclinB1 via ERCC1 to promote cell cycle progression.
ZNF326 promotes CyclinB1 protein synthesis via ERCC1. a H1299 cells and A549 cells were transfected with ZNF326-WT, ZNF326-ΔTAD, ZNF326-ΔZn, or control plasmid, respectively. The changes in CyclinB1 protein level and ERCC1 protein level were consistent. b Co-transfection of ZNF326 and ERCC1 in H1299 cells and A549 cells had a stronger effect on the upregulation of CyclinB1. c When H1299 cells and A549 cells were transfected with ZNF326 plasmids in conjunction with ERCC1 interference, this synergistic effect disappeared. d–g ERCC1 and ZNF326 both promoted colony formation and exhibited synergistic effects. Triplicate samples were analyzed for each treatment, and the results are presented as the mean ± SD, *p < 0.05, **p < 0.01
Discussion
The molecular structure of ZNF326 suggests that it functions as a transcription factor. We screened eight potential target genes by ChIP-seq and confirmed that ERCC1 is a target gene of ZNF326 using RT-PCR and dual-luciferase assays. Analyses with EMSA further confirmed that the sequence of region −883 to −875 bp in ERCC1 is the binding site of ZNF326. These results confirmed that ERCC1 is the target gene of ZNF326, and that ZNF326 initiates the transcriptional activity of ERCC1 via the binding of its C2H2 structure to the sequence of region −883 to −875 bp in the ERCC1 promoter. This process depends on the transcriptional activation domain of ZNF326. Our immunohistochemical analysis also supported the conclusion that ZNF326 upregulates the expression of ERCC1. Furthermore, there was a positive correlation between ZNF326 and ERCC1 expression in the 146 cases of NSCLC. However, the specificity of ERCC1 antibody of different clones has been controversial in recent studies. We used the FL-297 ERCC1 antibody to further verify the specificity of the 8F1 ERCC1 antibody [14,15,16]. We found that the FL-297 ERCC1 antibody mainly shojwed nuclear and cytoplasmic reaction in NSCLC tissues, whereas the 8F1 antibody showed cell nuclear reaction in NSCLC tissues (Supplementary Fig. 1A). The consistency rate of the two antibodies was 78.3% (Supplementary Table S7). In western blotting of the H1299 and A549 cell lines, the rabbit polyclonal FL-297 antibody against ERCC1 produced two bands at 37 kDa, which also appeared in the extracts. The mouse monoclonal 8F1 antibody against ERCC1 produced one band at 37 kDa for both cell lines (Supplementary Figure 1B). Thus, the 8F1 ERCC1 antibody was selected for immunohistochemistry and western blotting in our study. We further verified these results in subsequent experiments. After transfection of ERCC1 plasmid, Myc-tag was used for comparison. The position of the Myc-tag was consistent with that of 8F1, which was also verified in the ERCC1 interference experiment.
ERCC1 is a key factor in the process of nucleotide excision and repair; it mainly affects DNA replication, and changes in its RNA and protein levels can affect cell cycle progression [17, 18]. A previous study, using control cells and cells treated with cytotoxic agents, showed increased ERCC1 levels from the G1 through the S and G2 phases of the cell cycle in vitro [15]. Núñez et al. [19] showed that ERCC1-deficient hepatocytes were arrested in G2 and that DNA replication and the normal process of binucleation were both reduced. Similarly, CyclinB1 is also a chief cell cycle factor for the G2-to-M phase transition [20, 21]. Relevant studies showed that upregulation of CyclinB1 initiated the progression of cells from the G2 phase to the M phase and promoted mitosis and cell cycle arrest by downregulating CyclinB1 [22, 23]. Herein, we have shown that ZNF326 upregulated CyclinB1 protein level through ERCC1 to promote cell transition from G2 to M.
Our results suggested that ZNF326 promotes the growth of NSCLC cells. In contrast, Rangel et al. [6] proposed that ZNF326 is a tumor suppressor gene in triple-negative breast cancer. This discrepancy may be explained by the difference in the tumor cell lines used, and it is also possible that the target genes regulated by ZNF326 in various cell lines differ. In addition, the difference may be related to the antibodies, the sample size, and other factors.
Here we found that ZNF326 not only affects the cell cycle and cell proliferation but also changes the protein levels of E-cadherin, Snail, and Slug as well as the migration and invasion ability of lung cancer cells (Supplementary Fig. 2A-2D). These results suggested that ZNF326 is related to cell migration, invasion, and EMT, but that these changes are not mediated by ERCC1 (Supplementary Fig. 1E). In addition, ZNF326 may regulate multiple target genes, of which some are involved in transcriptional activation and others in transcriptional suppression. Future studies should go beyond the eight predicted target genes considered in this study.
In conclusion, ZNF326 plays a role as a tumor promoter in NSCLC, and its high expression correlates with low tumor differentiation and a high TNM stage. The C2H2 structure of ZNF326 can combine with region −883 to −875 bp of the ERCC1 promoter to promote ERCC1 transcription and translation and upregulate CyclinB1, which promotes cell cycle progression, thereby promoting the proliferation of NSCLC cells.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics 2012. CA Cancer J Clin. 2015;65:87–108.
Torre LA, Siegel RL, Ward EM, et al. Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiol Biomark Prev. 2016;25:16–27.
Chen WQ, Zuo TT, Zheng RS, et al. Lung cancer incidence and mortality in China in 2013. Zhonghua Zhong Liu Za Zhi. 2017;39:795–800.
Jen J, Lin LL, Chen HT, et al. Oncoprotein ZNF322A transcriptionally deregulates alpha-adducin, cyclin D1 and p53 to promote tumor growth and metastasis in lung cancer. Oncogene. 2016;35:2357–69.
Li K, Zhu ZC, Liu YJ, et al. ZFX knockdown inhibits growth and migration of non-small cell lung carcinoma cell line H1299. Int J Clin Exp Pathol. 2013;6:2460–7.
Rangel R, Guzman-Rojas L, Kodama T, et al. Identification of new tumor suppressor genes in triple-negative breast cancer. Cancer Res. 2017;7:4089–101.
Rengasamy M, Zhang F, Vashisht A, et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017;45:11106–20.
Close P, East P, Dirac-Svejstrup AB, et al. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484:386–9.
Lee JY, Kambe M, Hayashi M, et al. Cloning and characterization of a novel zinc finger protein that associates with nuclear matrix. DNA Cell Biol. 1998;17:849–58.
Travis WD, Brambilla E, Burke AP, et al. WHO classification of tumours of the lung, pleura, thymus, and heart. 4th ed. Lyon: IARC; 2015. p. 10–5.
Yang L, Wang S, Zhou Y, et al. Evaluation of the 7(th) and 8(th) editions of the AJCC/UICC TNM staging systems for lung cancer in a large North American cohort. Oncotarget. 2017;40:66784–95.
Andrieux LO, Fautrel A, Bessard A, et al. GATA-1 is essential in EGF-mediated induction of nucleotide excision repair activity and ERCC1 expression through ERK2 in human hepatoma cells. Cancer Res. 2007;67:2114–23.
Lee JY, Nakane Y, Koshikawa N, et al. Characterization of a zinc finger protein ZAN75: nuclear localization signal, transcriptional activator activity, and expression during neuronal differentiation of P19 cells. DNA Cell Biol. 2000;19:227–34.
Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. N Engl J Med. 2007;356:2538–40.
Dudás J, Schartinger VH, Romani A, et al. Cell cycle association and hypoxia regulation of excision repair cross complementation group 1 protein (ERCC1) in tumor cells of head and neck cancer. Tumour Biol. 2014;35:7807–19.
Vaezi AE, Bepler G, Bhagwat NR, et al. Choline phosphate cytidylyltransferase-α is a novel antigen detected by the anti-ERCC1 antibody 8F1 with biomarker value in patients with lung and head and neck squamous cell carcinomas. Cancer. 2014;120:1898–907.
Pérez-Ramírez C, Cañadas-Garre M, Alnatsha A, et al. Impact of DNA repair, folate and glutathione gene polymorphisms on risk of non small cell lung cancer. Pathol Res Pract. 2018;214:44–52.
Lindsey-Boltz LA. Bringing it all together: coupling excision repair to the DNA damage checkpoint. Photochem Photobiol. 2017;93:238–44.
Núñez F, Chipchase MD, Clarke AR, et al. Nucleotide excision repair gene (ERCC1) deficiency causes G(2) arrest in hepatocytes and a reduction in liver binucleation: the role of p53 and p21. FASEB J. 2000;14:1073–82.
Gao SY, Li J, Qu XY, et al. Downregulation of Cdk1 and cyclinB1 expression contributes to oridonin-induced cell cycle arrest at G2/M phase and growth inhibition in SGC-7901 gastric cancer cells. Asian Pac J Cancer Prev. 2014;15:6437–41.
Feng W, Cai D, Zhang B, et al. Combination of HDAC inhibitor TSA and silibinin induces cell cycle arrest and apoptosis by targeting survivin and CyclinB1/Cdk1 in pancreatic cancer cells. Biomed Pharmacother. 2015;8:257–64.
Cheng YM, Tsai CC, Hsu YC. Sulforaphane, a dietary isothiocyanate, induces G2/M arrest in cervical cancer cells through CyclinB1 downregulation and GADD45β/CDC2 association. Int J Mol Sci. 2016;17:12.
Xia YQ, Wei XY, Li WL, et al. Curcumin analogue A501 induces G2/M arrest and apoptosis in non-small cell lung cancer cells. Asian Pac J Cancer Prev. 2014;15:6893–8.
Acknowledgements
The ZNF326 interference target sequence was donated by Professor Roberto Rangel at the Anderson Cancer Center, University of Texas. This study was supported by the National Natural Science Foundation of China (No. 81572854 and No. 81772489, Wang Enhua).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wu, J., Zhang, X., Han, Q. et al. ZNF326 promotes proliferation of non-small cell lung cancer cells by regulating ERCC1 expression. Lab Invest 99, 169–179 (2019). https://doi.org/10.1038/s41374-018-0148-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0148-y
This article is cited by
-
Single-cell DNA methylation sequencing reveals epigenetic alterations in mouse oocytes superovulated with different dosages of gonadotropins
Clinical Epigenetics (2020)
-
ZNF326 promotes malignant phenotype of glioma by up-regulating HDAC7 expression and activating Wnt pathway
Journal of Experimental & Clinical Cancer Research (2019)