Abstract
Bronchial epithelial cells serve as a physical barrier at the forefront of the immune system. Barrier disruption and an excessive immune response of the bronchial epithelium contribute to the pathophysiology of asthma, a chronic bronchial inflammatory disease. The purpose of this study was to investigate the functional significance of ΔNp63, a p53-like transcription factor expressed by the basal bronchial epithelium. The immunohistochemical expression profile of ΔNp63 was evaluated in human bronchial tissue derived from asthma patients. The role of ΔNp63 in apoptosis inhibition and production of soluble mediators was investigated in vitro with cultured BEAS-2B bronchial epithelial cells using molecular biological analysis. In healthy bronchial tissue, ΔNp63-positive basal epithelial cells were covered with differentiated ΔNp63-negative cells but in the asthmatic airway, ΔNp63-positive cells were directly exposed to the bronchial lumen due to severe epithelial shedding. ΔNp63 regulated bronchial apoptosis in response to Toll-like receptor 3 stimulation. On the other hand, expression of ΔNp63 was modulated by stimulation with trypsin and SLIGKV, protease-activated receptor 2 ligands. Further phenotypic analysis revealed that ΔNp63 controlled the transcriptional expression and protein release of some epithelium-derived proinflammatory cytokines and endogenous protease inhibitors. We conclude that ΔNp63 modulates the bronchial epithelial response to viral infection. At the same time, ΔNp63 expression is influenced by proteases, which are abundant in house dust mites. Therefore, the ΔNp63 axis would be intimately involved in these two major triggers of asthma exacerbations, viral infection and protease overload.
Similar content being viewed by others
Introduction
Death from asthma has markedly decreased in the past decade due to better understanding of the disease and the development of improved therapeutic interventions [1], However, numerous children and adults still suffer from bronchial asthma, disrupting their quality of life. This occurs not only in countries that have long adopted a Western life style, but also in recently Westernized countries, with increasing numbers of patients reported. Thus, the socioeconomic burden attributed to asthma due to medical costs and loss of labor or education is now a worldwide problem. The growing prevalence of asthma patients along with lifestyle alterations in recent generations indicates that environmental factors are key to the epidemiology of asthma [2].
Bronchial asthma is a chronic inflammatory disease involving both immune cells and tissue cells [3, 4]. In particular, bronchial epithelial cells physically prevent exogenous particles from penetrating into subepithelial tissue and sense foreign antigens at the front line of the immune system. According to our current understanding, destruction of the tight junction barrier and excessive immune activation are fundamental epithelial alterations in the pathogenesis of bronchial asthma. Damaged or activated bronchial epithelial cells release various types of cytokines, including IL-33 and thymic stromal lymphopoietin (TSLP), which induce type 2 innate lymphoid cell and/or lymphocyte-mediated eosinophilic inflammation. On the other hand, type 2 inflammation-related cytokines affect bronchial epithelial integrity [5, 6]. This vicious cycle is currently the most persuasive explanation of asthma chronicity. However, although type 2 inflammation is an indispensable mechanism in asthma, asthma itself is not a single disease, but a syndrome that consists of numerous endotypes. In the early studies, histopathological analysis of asthmatic bronchial tissue showed severe eosinophil infiltration, a thickened basement membrane, submucosal fibrosis, smooth muscle hypertrophy, and distinctive features of epithelial lesions [7]. Such lesions included epithelial hyperplasia producing abundant mucus and epithelial shedding. It has been debated whether the latter finding is a true pathological finding or an artefact of the biopsy procedure [8, 9]. It is worth mentioning that basal epithelial cells remain even in damaged epithelium.
We previously investigated the functional significance of p63 and p73, members of the p53 family, in keratinocytes of patients with atopic dermatitis [10,11,12]. These proteins share highly conserved sequences and help to determine proliferation, apoptosis and differentiation in the context of carcinogenesis or development [13]. There are two major isoforms of p63: TAp63 and ΔNp63. While TAp63 is transcribed with the N-terminal transactivation domain from the P1 promoter, ΔNp63 lacks a transactivation domain. Nonetheless, ΔNp63 has transcriptional activities via an alternative P2 promoter [14]. Increasing numbers of studies have shown that ΔNp63, the predominant isoform of p63 expressed in epidermal keratinocytes and basal respiratory cells, modulates the expression of barrier-related proteins and immune reactions [15,16,17]. In addition, the expression of ΔNp63 is altered upon innate immune signaling [10, 11]. Therefore, we hypothesized that fine-tuning of ΔNp63 expression in epithelial cells would be intimately involved in the creation of the allergic tissue microenvironment. In this report, we show that ΔNp63-positive basal bronchial epithelial cells are resistant to the apoptosis induced by the Toll-like receptor 3 (TLR3) signal provided by viral infection. In the setting of epithelial shedding, the remaining ΔNp63-positive epithelial cells were exposed to the outer environment and revealed distinctive secretory activity of cytokines and anti-protease proteins.
Materials and Methods
Tissues
Bronchial tissues were obtained at autopsy from patients examined at Sapporo Medical University Hospital. The asthmatic donors were a 2-year-old female (Case 1), 79-year-old male (Case 2), and 26-year-old male (Case 3). The non-asthmatic control donor was a 68-year-old female. All tissues were obtained with written informed consent according to the guidelines of the Declaration of Helsinki and with approval of the Institutional Review Board of Sapporo Medical University Hospital under permit number 292–126, entitled Investigation of Human Diseases Utilizing Autopsy Specimen.
Cell cultures and stimulation
Human BEAS-2B bronchial epithelial cells were purchased from American Type Culture Collection (Manassas, VA). The cells were cultured as a monolayer in bronchial epithelial cell basal medium supplemented with the SingleQuots Kit (Lonza, Basel, Switzerland) at 37 °C in a humidified atmosphere with 5% CO2.
For cell stimulation, the culture medium was supplemented with polyinosine-polycytidylic acid (poly (I:C); Novus Biologicals, Littleton, CO), R837 (Novus Biologicals), ODN2006 (Hokkaido System Science, Sapporo, Japan), IFNγ (PeproTech, London, UK), IL-13 (PeproTech), trypsin (WAKO Pure Chemical, Osaka, Japan), papain (WAKO Pure Chemical), NH2-SLIGKV (Abcam, Cambridge, UK), and YM-58483 (Abcam) as indicated. The concentration of each reagent used is described in the figures or figure legends.
siRNA transfection
Human ΔNp63-specific small interfering RNA (siRNA) was purchased from Invitrogen (Carlsbad, CA; sense: 5′-ACAAUGCCCAGACUCAAUU-3′; antisense: 5′-AAUUGAGUCUGGGCAUUGU-3′). Scrambled siRNA for negative control was obtained from Invitrogen. Human TLR3-specific siRNA (sense: 5′-GAACUGGAUAUCUUUGCCATT-3′; antisense: 5′-UGGCAAAGAU AUCCAGUUCTT-3′) and control siRNA were purchased from Qiagen (Hilden, Germany). Transfections were carried out using Lipofectamine RNAiMAX (Invitrogen) in Opti-MEM (GIBCO, Carlsbad, CA) at 40 nmol/L according to the manufacturer’s instructions. The culture medium was replaced 6 h after transfection.
cDNA microarray analysis
mRNA was extracted from BEAS-2B bronchial epithelial cells transfected with ΔNp63-specific or scrambled siRNA 72 h after transfection. Microarray slides were scanned by a 3D-GENE human 25k (TORAY, Tokyo, Japan) and microarray images were automatically examined using AROSTM, version 4.0 (Operon Biotechnologies, Tokyo, Japan).
Antibodies
The antibodies used were a mouse monoclonal antibody to detect p63 (4A4; Abcam), mouse anti-ΔNp63 monoclonal antibody (clone; BC28, Biocare Medical, Pacheco, CA), mouse anti-α-tubulin monoclonal antibody (10G10; WAKO Pure Chemical), rabbit anti-cleaved poly (ADP-ribose) polymerase (PARP) monoclonal antibody (D64E10; Cell Signaling, Danvers, MA) and mouse anti-caspase 8 (1C12; Cell Signaling). Alexa 488 (green)-conjugated anti-mouse IgG was purchased from Invitrogen. Peroxidase-conjugated goat anti-mouse and anti-rabbit IgGs were obtained from KPL (Gaithersburg, MD).
PCR
Total RNA was extracted and purified using an RNeasy Mini Kit (Qiagen) and RNase-free DNase (Qiagen) according to the manufacturer’s instructions. Total RNA was reverse-transcribed into cDNA using a RevertAid RT kit containing random hexamers (Thermo Fisher Scientific, Woburn, MA). Quantitative PCR was performed with target gene-specific primers (Sigma-Aldrich, St. Louis, MO) and SYBR green PCR Master Mix (Thermo Fisher Scientific) on a StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA) as described in the manufacturer’s protocol. The sequences of the primers used to detect target gene expression are listed in Table S1. Elongation factor 1α (EF1α) mRNA was used to standardize the quantities of each transcript. To calculate the relative mRNA expression of triplicate specimens, the ΔΔCT method was used.
Immunohistochemical analysis
Sections (4 μm thick) of formalin-fixed paraffin-embedded tumors were immunostained using monoclonal antibodies after epitope retrieval with Target Retrieval Solution pH 9 (DAKO, Glostrup, Denmark). To detect ΔNp63 protein, the antibody was used at 1:100. Slides were counterstained with hematoxylin, rinsed, dehydrated in graded ethanol into non-aqueous solution, and then coverslipped with mounting media.
For fluorescent staining, cultured cells were fixed with 4% paraformaldehyde (WAKO Pure Chemical) and permeabilized with PBS containing 0.1% Triton X-100 (Sigma-Aldrich). They were incubated with the optimally diluted antibody at room temperature for 1 h and Alexa Fluor 488-labeled goat anti-mouse antibody (Invitrogen) under the same conditions for another 1 h. Then, slides were mounted with ProLong Gold Antifade Reagent (Invitrogen) containing 4’,6-diamidino-2-phenylindole (DAPI) for counterstaining of cell nuclei. Specimens were examined under an immunofluorescence microscope (IX81; Olympus, Tokyo, Japan).
Western blot analysis
Cells were lysed with RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA) containing protease inhibitors (Roche, Basel, Switzerland) for 30 min at 4 °C. Aliquots of the supernatants were applied to 5–20% SDS-PAGE under reducing conditions and transferred onto polyvinylidene fluoride membranes (Millipore, Bedford, MA). The membranes were incubated with blocking buffer and then with optimally diluted antibodies overnight at 4 °C. After being washed with wash buffer (0.05% Tween-20 in PBS), the membranes were reacted with a peroxidase-labeled secondary antibody for 1 h. After three washes with wash buffer, signals were visualized using an enhanced chemiluminescence detection system (Amersham Life Science, Arlington Height, IL). The intensity of the signals detected in the immunoblots was quantified using NIH ImageJ software. The intensity levels were normalized to the corresponding levels of α-tubulin.
Enzyme-linked immunosorbent assay
Culture supernatants were collected 72 h after experiment initiation and analyzed in triplicate to investigate protein concentrations with the Human DUOset ELISA kit (R&D Systems, Minneapolis, MN) for IL-1β, IL-8, and α1-antitrypsin, the Human Quantikine ELISA kit for secretory leukocyte proteinase inhibitor (SLPI), and the CircuLex Human ELISA Kit for α1-antichymotrypsin (Medical & Biological Laboratories, Nagoya, Japan) according to the manufacturers’ protocols. The absorbance of each sample was measured using an iMark microplate absorbance reader (Bio-Rad, Hercules, CA).
Cell viability assay
To investigate cell viability, WST-8 (Cell Counting Kit-8, Dojindo, Kumamoto, Japan) was added to each sample and incubated for 2 h at 37 °C in a CO2 incubator. The absorbance of each sample was measured using an iMark microplate absorbance reader.
Statistical analysis
Data analysis was performed with Prism Version 6 software (GraphPad Software, La Jolla, CA). Statistical significance was evaluated using the two-tailed unpaired Student’s t test or ANOVA with Tukey’s post-hoc tests. Graph bars in the figures indicate the mean ± SD. P values <0.05:* and <0.01:** are indicated inside the graphs. Each set of results shown is representative of at least three separate experiments.
Results
ΔNp63 affects bronchial epithelial survival during TLR3 stimulation
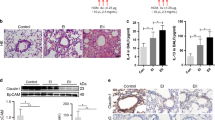
We examined the histopathology of the lung tissues derived from patients who died from an asthma attack. All had severe sputum embolism, basement membrane thickening, and eosinophil infiltration into the peribronchial submucosa. There were two types of epithelial lesions: epithelial hyperplasia (black arrow) and shedding (open arrow) (Fig. S1A). To investigate ΔNp63 expression in the bronchial epithelium, immunohistochemistry was performed with an antibody against ΔNp63. In non-asthmatic bronchial tissue, ΔNp63 was expressed in basal cells, which were covered with ΔNp63-negative ciliated columnar cells (Fig. 1a, upper panel). In asthmatic bronchial epithelium, despite severe epithelial shedding, there were still mixed population of ΔNp63-positive (high and low) basal cells (Fig. 1a, lower panel, Fig. S1B and S1C).
ΔNp63-positive basal bronchial epithelial cells are resistant to TLR3-induced apoptosis. a Hematoxylin and eosin (HE) staining (left panels) and immunohistochemistry for ΔNp63 (right panels) in non-asthmatic and asthmatic (case 1) bronchial tissue. Bar = 50 μm. b Relative cell viability between ΔNp63-knockdown and control BEAS-2B bronchial epithelial cells upon stimulation with representative TLR ligands mimicking viral infections. Cell viability was investigated 12 h after stimulation with 50 μg/mL poly (I:C), 50 μg/mL R837, or 10 μM ODN2006. n = 3. c Relative cell viability between TLR3 knockdown and control bronchial epithelial cells stimulated with 50 μg/mL poly (I:C) or 50 μg/mL poly (A:U). n = 3. d Expression of cleaved caspase 8 and cleaved PARP in ΔNp63-knockdown and control BEAS-2B bronchial epithelial cells with or without 4 h of 50 μg/mL poly (I:C) stimulation. **P < 0.01
Because viral infection is a major causative event in asthma exacerbations in both adults and children [18], we examined whether ΔNp63 expression affected epithelial survival in response to various types of Toll-like receptor stimulations mimicking viral infection. BEAS-2B human bronchial epithelial cells expressing abundant ΔNp63 were transfected with ΔNp63-specific siRNA, which successfully reduced ΔNp63 expression at transcript and protein levels (Fig. S2A and S2B). Poly (I:C) stimulation significantly reduced the viability of ΔNp63-knockdown bronchial epithelial cells when compared with control bronchial epithelial cells, whereas there was no difference in cell survival between the ΔNp63-knockdown and control groups under R837 (TLR7 ligand) and CpG-ODN (TLR9 ligand) stimulations (Fig. 1b). Because poly (I:C) might stimulate other double-stranded (ds) RNA receptors, we performed TLR3 knockdown using siRNA (data not shown). TLR3 knockdown diminished the cell death induced by poly (I:C) and poly (A:U), another TLR3 ligand (Fig. 1c). To investigate the mechanism of this TLR3-mediated decrease in cell viability, we examined the expression of apoptosis-related proteins via western blot analysis. As expected, downregulation of ΔNp63 increased cleaved caspase-8 and cleaved PARP levels in response to poly (I:C) stimulation (Fig. 1d), suggesting that ΔNp63 protects bronchial epithelial cells from viral dsRNA-mediated apoptosis.
Bronchial epithelial ΔNp63 levels are decreased by trypsin treatment
We next investigated which factors affect ΔNp63 expression in the bronchial epithelium. Type 2 innate lymphoid cells and helper T cells, crucial for the pathogenesis of asthma, produce abundant IL-13, which did not influence the ΔNp63 level; similar results were found for IFNγ, a representative type 1 inflammatory cytokine linked to airway hyperresponsiveness. In addition, viral infection-mimicking TLR ligands did not alter the ΔNp63 expression (Fig. S3A and S3B). Because some proteases derived from house dust mites affect epithelial functions, including physical barrier and cytokine release, and participate in the development of bronchial asthma [19], the bronchial epithelium was stimulated with these proteases. Interestingly, stimulation with trypsin, a serine protease, decreased the transcriptional and protein levels of ΔNp63 in a dose-dependent manner (Fig. 2a–c). On the other hand, papain, a cysteine protease, did not affect ΔNp63 expression, although papain stimulation did induce IL-6 expression (Fig. 2d, e, Fig. S4). As expected from the above results, cell viability was significantly decreased in the bronchial epithelial cells treated with trypsin and poly (I:C) (Fig. 2f).
ΔNp63 expression in BEAS-2B bronchial epithelial cells in response to serine and cysteine protease stimulation. a Transcriptional expression of ΔNp63 in bronchial epithelial cells in response to 24-h stimulation with 1, 10, and 100 nM trypsin, a serine protease. b Western blot analysis of p63 in bronchial epithelial cells in response to 48-h stimulation with 1, 10, and 100 nM trypsin. For western blotting, 4A4 antibody for pan-p63 was used. A single band reflecting ΔNp63, the dominant isoform in bronchial epithelium, was detected. c Fluor-labeled immunostaining for ΔNp63 after 48-h stimulation with 10 nM trypsin. Bar = 50 μm. d Transcriptional expression of ΔNp63 in bronchial epithelial cells in response to 24-h stimulation with 10, 100, and 1000 nM papain, a cysteine protease. e Western blot analysis of p63 in bronchial epithelial cells in response to 48-h stimulation with 10, 100, and 1000 nM papain. f Cell viability was measured in bronchial epithelial cells after 36-h stimulation with 10 nM trypsin or 100 nM papain and then 12-h stimulation with 50 μg/mL poly (I:C). n = 3. Numerical data indicate the relative intensity of the bands corrected by the corresponding levels of α-tubulin determined by ImageJ software. *P < 0.05 and **P < 0.01
Bronchial epithelial ΔNp63 is regulated by distinct trypsin pathways
Both trypsin and papain are proteases that can decrease cellular adhesion. In addition, they also induce signal transduction via a protease receptor. Because only trypsin decreased ΔNp63 levels, we hypothesized that receptor-mediated trypsin-sensing affects ΔNp63 expression. Thus, bronchial epithelial cells were treated with SLIGKV-NH2 peptide, an artificial ligand for protease-activated receptor-2 (PAR2), which behaved like trypsin and decreased transcriptional and protein levels of ΔNp63 (Fig. 3a, b). Because previous reports showed that SLIGKV-NH2 stimulation induced Ca2+ release-activated Ca2+ (CRAC) channel activation and thereby the production of the proinflammatory cytokines IL-6 and TSLP [18, 20], we pretreated bronchial epithelial cells with YM58483, a CRAC inhibitor. However, YM58483 did not block the decrease in ΔNp63, indicating that ΔNp63 regulation by trypsin is mediated by a different pathway from that of cytokine production.
Differential PAR2 signaling affects ΔNp63 expression in BEAS-2B bronchial epithelial cells. a Transcriptional expression of ΔNp63 in bronchial epithelial cells in response to 24-h stimulation with 5, 50, and 500 μM SLIGKV-NH2, a trypsin receptor agonist. b Western blot analysis of p63 in bronchial epithelial cells in response to 48-h stimulation with 5, 50, and 500 μM SLIGKV-NH2. c, d Bronchial epithelial cells were treated with 500 nM YM58483, a CRAC channel inhibitor, for 24 h. Consequently, 100 nM trypsin was added and the cells were harvested at 24 h for quantitative PCR or at 48 h for western blot. Then, the transcription levels of ΔNp63 (c) and protein expression levels of p63 (d) were investigated. Numerical data indicate the relative intensity of the bands corrected by the corresponding levels of α-tubulin determined by ImageJ software. *P < 0.05 and **P < 0.01
The expression profiles of bronchial epithelial cells regulated by ΔNp63
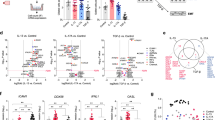
To investigate the transcriptional target(s) of ΔNp63 in BEAS-2B bronchial epithelial cells, bronchial epithelial cells transfected with ΔNp63-specific or control siRNA were subjected to cDNA microarray expression analysis. As shown in Tables S1 and S2, ΔNp63 modulated the expression levels of approximately 400 transcripts by more than 2-fold. Interestingly, ΔNp63 positively regulated IL1B, which encodes IL-1β, a pleiotropic proinflammatory cytokine. On the other hand, ΔNp63 negatively controlled the endogenous protease inhibitors SLPI and serine protease inhibitor A3 (SERPINA3; encoding α1-antichymotrypsin). We then investigated and confirmed the expression of these genes by quantitative PCR (Fig. 4a). In addition, transcriptional expression of IL-8 and serine protease inhibitor A1 (SERPINA1; encoding α1-antitrypsin) were also modulated by ΔNp63; this finding was not detected in the microarray analysis (Fig. 4a). As expected, the levels of IL-8, SLPI, α1-antichymotrypsin, and α1-antitrypsin protein in the culture supernatant of ΔNp63-knockdown bronchial epithelial cells were consistent with the gene expression levels. However, IL-1β was not detected at the protein level under conditions without any other stimulation, probably because a second signal is necessary for IL-1β release to activate the inflammasome that cleaves pro-IL-1β into mature IL-1β [21].
Modulation of cytokines and endogenous protease inhibitors by ΔNp63 in BEAS-2B bronchial epithelial cells. a Transcriptional expression of IL1B, IL8, SLPI, SERPINA3, and SERPINA1 in ΔNp63-knockdown and control bronchial epithelial cells. Cells were harvested 72 h after transfection. b Protein levels of endogenous IL-1β, IL-8, SLPI, α1-antichymotrypsin, and α1-antitrypsin in the culture supernatant of ΔNp63-siRNA-transfected and control bronchial epithelial cells. IL-1β protein was not detected without so-called second signals. n = 3. u.d., undetectable. **P < 0.01
The ΔNp63 level affects IL-1β and IL-8 production induced by poly (I:C) stimulation
As shown above, ΔNp63-positive bronchial epithelium was directly exposed in the setting of epithelial shedding. Here, we investigated whether there was a differential response of the bronchial epithelium between model of ΔNp63-positive basal and ΔNp63-negative apical epithelium using gene knockdown by ΔNp63-siRNA upon poly (I:C) stimulation mimicking viral infection. Consistent with the results in Fig. 4, control bronchial epithelium expressed significantly higher levels of IL1B and IL8 in a time- and poly (I:C) dose-dependent manner when compared to ΔNp63-knockdown cells (Fig. 5a, b). In addition, ΔNp63-knockdown bronchial epithelial cells released lower levels of mature IL-1β and IL-8 protein in response to poly (I:C) treatment (Fig. 5c).
IL-1β and IL-8 production induced by poly (I:C) stimulation is modulated by the expression level of ΔNp63. a, b Comparison of IL-1B and IL-8 expression in ΔNp63-siRNA-transfected and control bronchial epithelial cells in response to poly (I:C) stimulation. a Time course and (b) evaluation with different poly (I:C) doses. Cells were stimulated 48 h after siRNA transfection. Bronchial cells were stimulated with 5 μg/mL poly (I:C) in a. Cells were harvested 24 h after initiation of poly (I:C) stimulation in b. c Protein levels of endogenous mature IL-1β and IL-8 in the culture supernatant of ΔNp63-siRNA-transfected and control bronchial epithelial cells in response to poly (I:C) stimulation. Culture medium was replaced immediately before initiation of poly (I:C) stimulation and the supernatant was collected 48 h later. n = 3.**P < 0.01
Discussion
In this study, we investigated the differential bronchial epithelial behavior regulated by ΔNp63, a p53-like transcription factor. A schematic diagram summarizing the results of this study is shown in Fig. 6.
Dualism of ΔNp63 in bronchial epithelial shedding: schematic diagram of the findings and conclusions of this study. ΔNp63-negative apical bronchial epithelial cells would be sensitive to apoptosis induced by TLR3 stimulation. On the other hand, mixed population of ΔNp63-positive (high and low) basal cells were remained. Certain types of proteases decreased the expression of ΔNp63 in human bronchial epithelial cells. Production of some cytokines and protease inhibitors was regulated by ΔNp63 in human bronchial epithelial cells. Collectively, while ΔNp63 in the bronchial epithelial cells contributes to cell survival, ΔNp63-positive cells would produce higher amount of proinflammatory cytokines and lower levels of protease inhibitors. These results suggest that repeated protease overload and viral infection result in exposure of ΔNp63-positive basal bronchial epithelium and chronic inflammation in asthma
Several early studies proposed that epithelial shedding would be a characteristic histological finding of asthmatic bronchi. In the clinical setting, viral infection often results in asthma exacerbation [18]. This clinical phenomenon is at least partly explained by TLR3 stimulation of the production of TSLP, an inflammatory cytokine pivotal in the pathogenesis of bronchial asthma [22]. We showed that differentiated ΔNp63-negative bronchial epithelial cells tend to undergo apoptosis upon TLR3 stimulation, possibly reflecting the previous histological observation of apoptotic epithelium assessed by means of TUNEL (TdT-mediated dUTP nick end-labeling) staining [23]. Together with the evidence showing that ΔNp63 facilitates cell survival and inhibits apoptosis in various types of epithelial cells [13], ΔNp63-positive basal cells would be resistant to apoptosis following viral infection. The surviving ΔNp63-positive basal bronchial epithelial cells are assumed to be reserve cells for restoring epithelial integrity.
In our observations, bronchial ΔNp63 expression was decreased by trypsin but not papain, suggesting that distinctive types of proteases induce different responses of epithelial cells. Excessive exposure to proteases is due to exogenous proteases, mainly from house dust mite, and endogenous proteases, including human airway trypsin-like protease (HAT) and HAT-like proteases [24]. These proteases are classically categorized according to cleavage site. Both trypsin (serine protease) and papain (cysteine protease), as well as their related proteases, induce PAR2-mediated epithelial cytokine production, although PAR2 provokes biased signal transduction depending on the type of protease [25]. In addition, CRAC channel inhibition, which blocks PAR2-mediated cytokine secretion, could not rescue ΔNp63 suppression, indicating the need for further elucidation of the PAR2 pathway.
Previous work showed that proteases decrease the tight junction barrier and thereby enhance the penetration of various extrinsic antigens, including those with trypsin-like protease activity [26]. Therefore, contact between trypsin-like protease and ΔNp63-positive basal cells would worsen epithelial shedding, which is mediated by epithelial apoptosis through viral TLR3 ligand signaling. In the setting of epithelial shedding, there were still some ΔNp63-positive cells, which are supposed to produce lower amounts of SLPI, α1-antichymotrypsin and α1-antitrypsin. Although the functional significance of insufficiency in these protease inhibitors in asthma remains to be fully elucidated, unfavorable deviations in proteases and their inhibitor production in the tissue microenvironment are considered to underlie the pathogenesis of the disease [19]. Indeed, increased prevalence of asthma is already known in patients with α1-antitrypsin deficiency [27]. In addition, a copy number variation of SERPINA3 (encoding α1-antichymotrypsin) may also be involved in asthma [28]. It is worth mentioning that anti-inflammatory and tissue protective functions of SLPI have been extensively studied in various organs [29]. Importantly, decreased bronchial SLPI is reported in severe human asthma [30].
Taken together with our findings, these results suggest that recurrent viral infection provokes sustained exposure of ΔNp63-positive basal cells with lower levels of endogenous protease inhibitors, resulting in the release of protease-induced asthmogenic cytokines, including TSLP, from epithelium [31, 32]. We found that ΔNp63 transcriptionally modulated the production of some cytokines. Neutrophils recruited by IL-8 are critical to the pathophysiology of severe asthma [33]. IL-1β is a pleiotropic cytokine that augments a wide range of effects on the immune system or tissue remodeling [34]. Notably, a recent study showed the critical role played by the IL-1β pathway in Th2/Th17-predominant type asthma [35]. Our data would indicate that exposed ΔNp63-positive bronchial epithelial cells release abundant amounts of these cytokines upon dsRNA stimulation in the context of viral infection, which would lead to neutrophilic inflammation and tissue remodeling, hallmarks of asthmatic bronchial tissue.
Significantly, the tissue showed signs of allergic inflammation, with epidermal keratinocyte, sinonasal, conjunctival, and esophageal epithelial cells strongly expressing ΔNp63, a master regulator of differentiation. We previously showed that ΔNp63 and ΔNp73, p53 family proteins, play important roles in the pathophysiology of atopic dermatitis [10,11,12]. In addition, a growing number of studies have shown that p63 and p73 are critical for the development of the tight junction barrier and/or ciliogenesis in respiratory epithelium [15, 36, 37]. Therefore, the non-oncogenic functions of p53 family proteins may play potentially fundamental roles in modulating the physical barrier and the immunological activity of the epithelium in allergic inflammations, besides the well-known biological role of p53 homologs in determining cell fate in the context of the development of carcinogenesis [13]. Furthermore, since ΔNp63 regulated expression of epithelial related genes, as listed in the Tables S1 and S2, ΔNp63 may be involved in epithelial-mesenchymal transition in inflammatory conditions [38]. Interestingly, in eosinophilic esophagitis, the esophageal epithelium expresses altered transcript levels of IL-1 family protein, some protease inhibitors and differentiation-related molecules, suggesting that fluctuations in ΔNp63 expression potentially underlie the disease [39].
In conclusion, we determined the potential functional relevance of ΔNp63 in epithelial shedding, a characteristic pathological finding of asthma. The expression of ΔNp63 was regulated by trypsin and SLIGKV, PAR2 ligands. On the other hand, ΔNp63 moderated the production of IL-1β, IL-8, SLPI, α1-antichymotrypsin, and α1-antitrypsin. Therefore, in the setting of epithelial shedding, exposed ΔNp63-positive bronchial epithelium should show reduced resistance to protease because of lower levels of protease inhibitors, which would result in a further acceleration of barrier disruption and excessive immune activation of the epithelium. Although we did not show the involvement of type 2 inflammation in this study, our results may link the two major exacerbating factors of asthma, viral infection and protease overload. We suggest that it is worth focusing on the distinctive behavior and response between stem-like basal and differentiated apical bronchial epithelial cells to reveal the functional role of epithelial cells in asthma. ΔNp63 would be an essential participant, although there should be other factors that are independent of ΔNp63. Additional investigation into non-tumorigenic but inflammation-related functions of the p53 family molecules together with other differentiation factors may lead to further understanding of the pathogenesis of the allergic disorders and their mechanism of chronicity.
References
Platts-Mills TA. The allergy epidemics: 1870-2010. J Allergy Clin Immunol. 2015;136:3–13.
Stein MM, Hrusch CL, Gozdz J, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;375:411–21.
Gon Y, Hashimoto S. Role of airway epithelial barrier dysfunction in pathogenesis of asthma. Allergol Int. 2018;67:12–17.
Loxham M, Davies DE. Phenotypic and genetic aspects of epithelial barrier function in asthmatic patients. J Allergy Clin Immunol. 2017;139:1736–51.
Sugita K, Steer CA, Martinez-Gonzalez I, et al. Type 2 innate lymphoid cells disrupt bronchial epithelial barrier integrity by targeting tight junctions through IL-13 in asthmatic patients. J Allergy Clin Immunol. 2018;141:300–310 e311.
Wawrzyniak P, Wawrzyniak M, Wanke K, et al. Regulation of bronchial epithelial barrier integrity by type 2 cytokines and histone deacetylases in asthmatic patients. J Allergy Clin Immunol. 2017;139:93–103.
Sumi Y, Hamid Q. Airway remodeling in asthma. Allergol Int. 2007;56:341–8.
Ordonez C, Ferrando R, Hyde DM, et al. Epithelial desquamation in asthma: artifact or pathology? Am J Respir Crit Care Med. 2000;162:2324–9.
Ordonez CL, Fahy JV. Epithelial desquamation in asthma. Am J Respir Crit Care Med. 2001;164(10 Pt 1):1997.
Kubo T, Kamekura R, Kumagai A, et al. DeltaNp63 controls a TLR3-mediated mechanism that abundantly provides thymic stromal lymphopoietin in atopic dermatitis. PLoS One. 2014;9:e105498.
Kubo T, Sugimoto K, Kojima T, et al. Tight junction protein claudin-4 is modulated via DeltaNp63 in human keratinocytes. Biochem Biophys Res Commun. 2014;455:205–11.
Kumagai A, Kubo T, Kawata K, et al. Keratinocytes in atopic dermatitis express abundant DeltaNp73 regulating thymic stromal lymphopoietin production via NF-kappaB. J Dermatol Sci. 2017;88:175–83.
Pflaum J, Schlosser S, Muller M. p53 Family and cellular stress responses in cancer. Front Oncol. 2014;4:285.
Allocati N, Di Ilio C, De Laurenzi V. p63/p73 in the control of cell cycle and cell death. Exp Cell Res. 2012;318:1285–90.
Kaneko Y, Kohno T, Kakuki T, et al. The role of transcriptional factor p63 in regulation of epithelial barrier and ciliogenesis of human nasal epithelial cells. Sci Rep. 2017;7:10935.
Kojima T, Kohno T, Kubo T, et al. Regulation of claudin-4 via p63 in human epithelial cells. Ann N Y Acad Sci. 2017;1405:25–31.
Rizzo JM, Oyelakin A, Min S, et al. DeltaNp63 regulates IL-33 and IL-31 signaling in atopic dermatitis. Cell Death Differ. 2016;23:1073–85.
Edwards MR, Strong K, Cameron A, et al. Viral infections in allergy and immunology: How allergic inflammation influences viral infections and illness. J Allergy Clin Immunol. 2017;140:909–20.
Reed CE, Kita H. The role of protease activation of inflammation in allergic respiratory diseases. J Allergy Clin Immunol. 2004;114:997–1008. quiz 1009
Jairaman A, Yamashita M, Schleimer RP, et al. Store-operated Ca2 + release-activated Ca2 + channels regulate PAR2-activated Ca2 + signaling and cytokine production in airway epithelial cells. J Immunol. 2015;195:2122–33.
Prochnicki T, Mangan MS, Latz E. Recent insights into the molecular mechanisms of the NLRP3 inflammasome activation. F1000Res. 2016;5:1469.
Kato A, Favoreto S Jr, Avila PC, et al. TLR3- and Th2 cytokine-dependent production of thymic stromal lymphopoietin in human airway epithelial cells. J Immunol. 2007;179:1080–7.
Trautmann A, Schmid-Grendelmeier P, Kruger K, et al. T cells and eosinophils cooperate in the induction of bronchial epithelial cell apoptosis in asthma. J Allergy Clin Immunol. 2002;109:329–37.
Menou A, Duitman J, Flajolet P, et al. Human airway trypsin-like protease, a serine protease involved in respiratory diseases. Am J Physiol Lung Cell Mol Physiol. 2017;312:L657–68.
Zhao P, Metcalf M, Bunnett NW. Biased signaling of protease-activated receptors. Front Endocrinol. 2014;5:67.
Kale SL, Agrawal K, Gaur SN, et al. Cockroach protease allergen induces allergic airway inflammation via epithelial cell activation. Sci Rep. 2017;7:42341.
Eden E, Strange C, Holladay B, et al. Asthma and allergy in alpha-1 antitrypsin deficiency. Respir Med. 2006;100:1384–91.
Rogers AJ, Chu JH, Darvishi K, et al. Copy number variation prevalence in known asthma genes and their impact on asthma susceptibility. Clin Exp Allergy. 2013;43:455–62.
Majchrzak-Gorecka M, Majewski P, Grygier B, et al. Secretory leukocyte protease inhibitor (SLPI), a multifunctional protein in the host defense response. Cytokine Growth Factor Rev. 2016;28:79–93.
Raundhal M, Morse C, Khare A, et al. High IFN-gamma and low SLPI mark severe asthma in mice and humans. J Clin Invest. 2015;125:3037–50.
Kouzaki H, O’Grady SM, Lawrence CB, et al. Proteases induce production of thymic stromal lymphopoietin by airway epithelial cells through protease-activated receptor-2. J Immunol. 2009;183:1427–34.
Matsumura Y. Role of allergen source-derived proteases in sensitization via airway epithelial cells. J Allergy. 2012;2012:903659.
Hosoki K, Itazawa T, Boldogh I, et al. Neutrophil recruitment by allergens contribute to allergic sensitization and allergic inflammation. Curr Opin Allergy Clin Immunol. 2016;16:45–50.
Pinkerton JW, Kim RY, Robertson AAB, et al. Inflammasomes in the lung. Mol Immunol. 2017;86:44–55.
Liu W, Liu S, Verma M, et al. Mechanism of TH2/TH17-predominant and neutrophilic TH2/TH17-low subtypes of asthma. J Allergy Clin Immunol. 2017;139:1548–58 e1544.
Nemajerova A, Kramer D, Siller SS, et al. TAp73 is a central transcriptional regulator of airway multiciliogenesis. Genes Dev. 2016;30:1300–12.
Jackson PK, Attardi LD. p73 and FoxJ1: programming multiciliated epithelia. Trends Cell Biol. 2016;26:239–40.
Stacy AJ, Craig MP, Sakaram S, et al. DeltaNp63alpha and microRNAs: leveraging the epithelial-mesenchymal transition. Oncotarget. 2017;8:2114–29.
Rochman M, Travers J, Miracle CE, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140:738–49 e733.
Acknowledgements
This research received a grant from the Kao Foundation for Arts and Sciences and a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS) to TK (Grant Number 16K21249).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kubo, T., Tsujiwaki, M., Hirohashi, Y. et al. Differential bronchial epithelial response regulated by ΔNp63: a functional understanding of the epithelial shedding found in asthma. Lab Invest 99, 158–168 (2019). https://doi.org/10.1038/s41374-018-0132-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0132-6