Abstract
Multiple myeloma (MM) commonly displays multidrug resistance and is associated with poor prognosis. Therefore, it is important to identify the mechanisms by which MM cells develop multidrug resistance. Our previous study showed that multidrug resistance is correlated with overexpression of multidrug resistance protein 1 (MDR1) and Survivin, and downregulation of Bim expression in melphalan-resistant RPMI8226/L-PAM cells; however, the underlying mechanism of multidrug resistance remains unclear. In the present study, we investigated the mechanism of multidrug resistance in melphalan-resistant cells. We found that RPMI8226/L-PAM and ARH-77/L-PAM cells showed increased phosphorylation of extracellular signal-regulated protein kinase 1/2 (ERK1/2) and Akt, and nuclear localization of nuclear factor κB (NF-κB). The combination of ERK1/2, Akt, and NF-κB inhibitors with melphalan reversed melphalan resistance via suppression of Survivin expression and enhanced Bim expression in melphalan-resistant cells. In addition, RPMI8226/L-PAM and ARH-77/L-PAM cells overexpressed hypoxia-inducible factor 1α (HIF-1α) via activation of ERK1/2, Akt, and NF-κB. Moreover, suppression of HIF-1α by echinomycin or HIF-1α siRNA resensitized RPMI8226/L-PAM cells to melphalan through downregulation of Survivin expression and upregulation of Bim expression. These results indicate that enhanced Survivin expression and decreased Bim expression by HIF-1α via activation of ERK1/2, Akt, and NF-κB play a critical role in melphalan resistance. Our findings suggest that HIF-1α, ERK1/2, Akt, and NF-κB inhibitors are potentially useful as anti-MDR agents for the treatment of melphalan-resistant MM.
Similar content being viewed by others
Introduction
Multiple myeloma (MM) is a malignancy of plasma cells. It is the second most common hematopoietic tumor and is characterized by expansion and accumulation of plasma cells in the bone marrow [1, 2]. The 5-year survival rate for multiple myeloma is estimated to be about 50.7%, on the basis of data from the Surveillance, Epidemiology, and End Results (SEER) program [3, 4]. Although the development of new therapeutic agents, including proteasome inhibitors, such as bortezomib, cafilzomib, and ixazomib, and immunomodulatory drugs, such as thalidomide and lenalidomide has improved the survival rate of patients with MM, MM remains incurable, and relapses are frequent, and decreased sensitivity to chemotherapy leads to disease progression in more than 80% of the patients [5,6,7,8,9,10,11]. Melphalan, an alkylating agent, in combination with predonisolone has been used as first-line therapy in patients with MM and has proven effective in MM therapy in non-transplant candidates. In addition, a combination of melphalan and predonisolone plus bortezomib or lenalidomaide/thalidomide prolongs overall survival in patients with MM [12,13,14,15]. Furthermore, a combination of dratumumab, bortezomib, melphalan, and predonisolone (daratumab group) increases the progression-free survival (PFS) of patients in 18 months relatively to bortezomib, melphalan, and predonisolone alone (control group), and decreases disease progression or death from 40.2 to 25.1% of patients [16]. However, chemotherapy regimens directed to MM patients with predicted resistance to melphalan and/or bortezomib do not lead to higher progression-free survival than those directed to patients with predicted sensitivity to melphalan and/or bortezomib [17]. MM cells frequently exhibit multidrug resistance (MDR) against common conventional cytotoxic cancer drugs, and therefore it is important to identify the MDR mechanism.
P-glycoprotein, the product of multidrug resistance protein 1 (MDR1) gene, is implicated as the one factor of MDR in tumors where it acts as an efflux pump. MDR1 is located in the transmembrane and reduces the intracellular concentration of anticancer drugs, thereby causing MDR [18, 19]. Bcl-2 family members regulate response to chemotherapy in various cancer cells [20]. Bim, the BH-3 only pro-apoptotic protein of Bcl-2 family, promotes apoptosis and reduces protein levels, leading to drug resistance [6, 21]. In addition, Survivin, a member of the inhibitor of apoptosis family, suppresses apoptosis via inhibition of caspase activity and is involved in acquired drug resistance [22,23,24]. These proteins are regulated by various signaling pathways. It has been reported that the phosphoinositide 3-kinase (PI3K)/Akt and mitogen-activated protein kinase (MAPK) kinase (MEK)/extracellular signal-regulated protein kinase 1/2 (ERK1/2) pathways induce overexpression of Bcl-2 and downregulation of Bim [25]. It has also been reported that MDR1-mediated MDR involves several signaling pathways, including PI3K/Akt, MEK/ERK, and nuclear factor κB (NF-κB) pathways [26, 27]. Moreover, activation of ERK1/2, Akt, and NF-κB induces the expression of Survivin [28,29,30]. Thus, modulation of these signaling pathways may overcome MDR in MM cells.
We previously used the myeloma cell line RPMI8226 and its multidrug-resistant counterpart RPMI8226/L-PAM to investigate MDR mechanisms, and found that MDR1 and Survivin are overexpressed, and Bim is suppressed in RPMI8226/L-PAM cells [6]. We also previously demonstrated that activation of c-Src contributes to the MDR observed in these cells [31]. Although the expression of MDR1, Survivin, and Bim is regulated by c-Src activation in RPMI8226/L-PAM cells, the underlying mechanism of MDR in MM cells remains unclear. In the present study, we further investigated the mechanism of melphalan resistance in melphalan-resistant cells.
Materials and methods
Materials
Melphalan, echinomycin, U0126, LY294002, and dimethyl fumarate (DMF) were purchased form Wako (Tokyo, Japan). These reagents were dissolved in dimethyl sulfoxide. These reagents were diluted in PBS (0.05 M, pH7.4), filtrated through syringe filters (0.45 μm, IWAKI GLASS, Tokyo, Japan) and used for various assays described below.
Cell culture
RPMI8226 cells were obtained from Health Science Research Resources Bank (Osaka, Japan). ARH-77 cells were obtained from DS Pharma Biomedical (Osaka, Japan). Melphalan-resistant variant were established in our laboratory. This resistant variants are resistant to adriamycin, vincristine, dexamethasone, and melphalan. These cells were cultured in RPMI1640 medium (Sigma) supplemented with 10% fetal calf serum (Gibco, Carlsbad, CA, USA), 100 μg/ml penicillin (Gibco), 100 U/ml streptomycin (Gibco), and 25 mM HEPES (pH 7.4; Wako) in an atmosphere containing 5% CO2.
Trypan blue dye exclusion assay
The effect of various drugs on cell survival/proliferation was determined using the trypan blue dye exclusion assay. Prior to each experiment, cells (2 × 103 cells/well) were plated onto 96-well plates. After culturing for 24 h, the cells were exposed to anticancer drugs for various times. Equal volumes of cell suspension and 0.4% trypan blue solution were mixed gently, loaded into a hemocytometer, and the viable cells (unstained) and dead cells (stained blue) were counted. Each experiment was performed in triplicate. Results are reported from an average of at least five independent experiments.
Western blotting
The cytoplasm and nuclear fraction of cells was extracted with the ProteoExtract Subcellular Proteome Extraction Kit (Calbiochem, San Diego, CA, USA). The protein content in the cell lysates was determined using a BCA protein-assay kit. The extracts (40 μg of protein) were fractionated on polyacrylamide-SDS gels and transferred to PVDF membranes (GE Healthcare, Buckinghamshire, UK). The membranes were blocked with a solution containing 3% skim milk and incubated overnight at 4 °C with each of the following antibodies: anti-NF-κB p65, anti-phospho-ERK1/2 antibody, anti-phospho-Akt antibody, anti-phospho-mammalian target of rapamycin (mTOR) antibody, anti-phospho-p38MAPK, anti-phospho-c-Jun N-terminal kinase (JNK), antibody, anti-ERK1/2 antibody, anti-Akt antibody, anti-mTOR antibody, anti-p38MAPK antibody, anti-HIF-1α antibody (Cell Signaling Technology, Beverly, MA, USA), anti-MDR antibody, anti-Survivin antibody, anti-Bim antibody, anti-lamin A/C antibody (Santa Cruz Biotechnologies, CA, USA), and anti-β-actin antibody (Sigma). Subsequently, the membranes were incubated with horseradish peroxidase-coupled anti-rabbit IgG sheep antibodies (GE Healthcare) for 1 h at room temperature. The reactive proteins were visualized using Luminata Forte (Merck Millipore, Nottingham, UK) according to the manufacturer’s instructions.
HIF-1α transcriptional activity
The DNA binding activity of HIF-1α was evaluated using HIF-1α transcription factor assay kit (Cayman Chemical, MI, USA) according to the manufacturer’s instructions. Nuclear extracts were prepared and incubated in 96-well plates coated with immobilized double-stranded oligonucleotides containing the HIF-1α response element. The HIF-1α transcription factor complex was detected by the addition of a specific primary antibody directed against HIF-1α, visualized by an anti-IgG horseradish peroxidase-conjugate and quantified by measuring the absorbance at 450 nm.
RNA interference
The double-stranded HIF-1α small interfering RNAs (siRNAs; HSS104774), MDR1 small interfering RNAs (siRNAs; HSS107919), Survivin siRNAs (HSS179403), and Bim siRNAs (HSS145413) were synthesized and purified by Invitrogen (Carlsbad, CA, USA). StealthTM RNAi negative control duplex (low GC content) (Invitrogen) was used as a negative control. Transfection of siRNAs was performed according to the manufacturer’s protocol by using the LipofectAMINETM 3000 reagent (Invitrogen). Briefly, 4 μl of 20-μM siRNA was mixed with 200 μl of Opti-MEM. LipofectAMINE TM 3000 (4 μl) was diluted in 200 μl of Opti-MEM and incubated at room temperature for 5 min. After incubation, the diluted LipofectAMINETM 3000 was mixed with the diluted siRNA and further incubated for 20 min at room temperature. Total 400 μl of the siRNA-LipofectAMINETM 3000 complex was applied to each well of the cultured cells at approximately 50–70% confluence in 6-well microplates.
Quantitative real-time polymerase chain reaction (PCR)
Total RNA was isolated using RNAiso (Takara Biomedical, Siga, Japan). One microgram of purified total RNA was used for the real-time PCR analysis with the PrimeScript First-Strand Synthesis System (Takara Biomedical). cDNA was subjected to quantitative real-time PCR by using SYBR Premix Ex Taq (Takara Biomedical) and the ABI Prism 7000 detection system (Applied Biosystems, Foster, CA) in a 96-well plate according to the manufacturer’s instructions. The PCR conditions for GAPDH, MDR1, Survivin, and Bim were 94 °C for 2 min; followed by 40 cycles of 94 °C for 0.5 min, 50 °C for 0.5 min, and 72 °C for 0.5 min. The following primers were used: MDR1, 5′-GGC TCC GAT ACA TGG TTT TCC-3′ (5′-primer) and 5′-TTC AGT GTG CGA TCT TCC CAG C-3′ (3′-primer); Survivin, 5′-CTG CCT GGC AGC CCT TTC TCA A-3′ (5′-primer) and 5′-AAT AAA CCC TGG AAG TGG TGC A-3′ (3′-primer); Bim, 5′-ATG GCA AAG CAA CCT TCT GA-3′ (5′-primer) and 5′-CGC ATA TCT GCA GGT TCA GCC-3′ (3′-primer); and GAPDH, 5′-GAC ATC AAG AAG GTG GTG AA-3′ (5′-primer) and 5′-TGT CAT ACC AGG AAA TGA GC-3′ (3′-primer). As an internal control for each sample, the GAPDH gene was used for standardization. Cycle threshold (Ct) values were established, and the relative difference in expression from GAPDH expression was determined according to the 2–∆∆Ct method of analysis and compared to the expression in control cells.
SurvExpress analysis
The relationship between HIF-1α expression levels and the overall survival of MM patients was evaluated by the P-value approach using the SurvExpress database [32]. Microarray datasets of Shaughnessy [33] were assessed using SurvExpress database. Briefly, patients were divided into two groups according to HIF-1α expression levels in their tumors, at all possible cut-off points.
Statistical analysis
All results are expressed as means and S.D. of several independent experiments. Multiple comparisons of the data were done by ANOVA with Dunnet’s test. P values less than 5% were regarded as significant.
Results
ERK1/2, Akt, and NF-κB are constitutively activated in melphalan-resistant cells
The results of our previous study indicated that MDR1 and Survivin expression is elevated, Bim expression is decreased, and c-Src is activated in the melphalan-resistant RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cell lines that we established (Supplementary Fig. S1 and S2) [31]. We therefore investigated the activity of the downstream effectors of Src in RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells. The expression level of phosphorylated ERK1/2 and Akt was higher in RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells than in RPMI8226, ARH-77/L-PAM, and HS-Sultan cells (Fig. 1 and Supplementary Fig. S3). Nuclear localization of NF-κB was elevated in RPMI8226/ L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells (Fig. 1 and Supplementary Fig. S3). However, the expression levels of phosphorylated mTOR, p38MAPK, and JNK did not differ between the parent and drug-resistant cells (Fig. 1 and Supplementary Fig. S3).
Melphalan-resistant cells express constitutively phosphorylated ERK1/2, Akt, and NF-κB. a The cytoplasmic and nuclear fractions were extracted from RPMI8226 and RPMI8226/L-PAM cells and then subjected to SDS-PAGE/immunoblotting with anti-phospho-ERK1/2, anti-phospho-mTOR, anti-phospho-p38MAPK, anti-phospho-Akt, anti-phospho-JNK, anti-NF-κB p65, anti-ERK1/2, anti-Akt, anti-mTOR, anti-JNK, and anti-p38MAPK antibodies. Anti-β-actin and anti-lamin antibodies were used as internal standards. b Quantification of the amount of NF-κB p65, phospho-ERK1/2, phospho-Akt, phospho-mTOR, or phospho-p38MAPK, normalized to the amount of the corresponding proteins, respectively. The results are representative of five independent experiments. *P < 0.01 compared to RPMI8226 cells (ANOVA with Dunnett’s test). c The cytoplasmic and nuclear fractions were extracted from ARH-77 and ARH-77/L-PAM cells and then subjected to SDS-PAGE/immunoblotting with anti-phospho-ERK1/2, anti-phospho-mTOR, anti-phospho-p38MAPK, anti-phospho-Akt, anti-phospho-JNK, anti-NF-κB p65, anti-ERK1/2, anti-Akt, anti-mTOR, anti-JNK, and anti-p38MAPK antibodies. Anti-β-actin and anti-lamin antibodies were used as internal standards. d Quantification of the amount of NF-κB p65, phospho-ERK1/2, phospho-Akt, phospho-mTOR or phospho-p38MAPK, normalized to the amount of the corresponding proteins, respectively. The results are representative of five independent experiments. *P < 0.01 compared to ARH-77 cells (ANOVA with Dunnett’s test)
Inhibition of ERK1/2, Akt, and NF-κB activation overcomes drug resistance in RPMI8226/L-PAM cells
The potential role of ERK1/2, Akt, and NF-κB in the drug resistance displayed by RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells was further investigated, and it was found that ERK1/2 and Akt phosphorylation, and the nuclear entry of NF-κB p65 were inhibited by treatment with 5 or 10 μM U0126 (a MEK inhibitor), 5 or 10 μM LY294002 (a PI3K inhibitor), and 20 μM DMF (an NF-κB inhibitor) in RPMI8226/L-PAM, ARH-77/L-PAM, or HS-Sultan/L-PAM cells, respectively (Fig. 2a–f, Supplementary Fig. S4a–c, and Fig. S5a–c). Next, the effect of melphalan alone and in combination with one, two, or all three of these inhibitors (U0126, LY294002, and DMF) on the viability of RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells was assessed. Each of the three inhibitors was shown to weakly enhance the sensitivity of RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells to melphalan (Fig. 2g, Supplementary Fig. S4d, and Fig. S5d). Double-inhibitor combinations were moderately enhanced the sensitivity of cells to melphalan, and triple-inhibitor combinations were found to significantly overcome drug resistance (Fig. 2g, Supplementary Fig. S4d, and Fig. S5d). The inhibitors alone were found to be non-toxic to these cells (Supplementary Fig. S6-S8).
Inhibition of ERK1/2, Akt, or NF-κB overcomes drug resistance in RPMI8226/L-PAM cells. a–f RPMI8226/L-PAM cells were exposed to the indicated concentrations of U0126, LY294002, or dimethyl fumarate (DMF). a–c After incubation for 72 h, the cytoplasmic and nuclear fractions were extracted and then subjected to SDS-PAGE/immunoblotting with anti-phospho-ERK1/2, anti-phospho-Akt, anti-ERK1/2, anti-Akt, and anti-NF-κB p65 antibodies. Anti-β-actin and anti-lamin antibodies were used as internal standards. d–f Quantification of the amount of NF-κB p65, phospho-ERK1/2, or phospho-Akt, normalized to the amount of the corresponding proteins, respectively. The results are representative of five independent experiments. *P < 0.01 compared to RPMI8226 cells (ANOVA with Dunnett’s test). g RPMI8226/L-PAM cells were exposed to the indicated concentrations of melphalan, U0126, LY294002, or DMF. After incubation for 72 h, the number of dead cells was counted by trypan blue staining. The results are representative of five independent experiments. *P < 0.01 vs. RPMI8226 untreated cells, #P < 0.01 vs. RPMI8226/L-PAM untreated cells (ANOVA with Dunnett’s test)
The effect of signaling molecule inhibitors on the expression of MDR1, Survivin, and Bim
The effects of U0126, LY294002, and DMF treatment on RPMI8226/L-PAM cells were further assessed in terms of the molecular mechanisms associated with MDR1, Survivin, and Bim expression. Treatment with U0126, LY294002, and DMF alone was found to reduce the expression of MDR1, but not that of Survivin, and did not increase Bim expression (Fig. 3a, d). Double-inhibitor combinations reduced the expression of Survivin and MDR1 to levels similar to those of parent cells, and weakly increased Bim expression (Fig. 3b, e). Triple-inhibitor combinations decreased MDR1 and Survivin expression, and enhanced Bim expression (Fig. 3c, f).
U0126, LY294002, and DMF treatment suppresses Survivin expression and enhances Bim expression in RPMI8226/L-PAM cells. a–c RPMI8226/L-PAM cells were exposed to the indicated concentrations of U0126, LY294002, or dimethyl fumarate (DMF). After incubation with inhibitors for 72 h, the cell lysates were extracted and subjected to SDS-PAGE/immunoblotting with antibodies against MDR1, Survivin, and β-actin. Anti-β-actin antibody was used as an internal standard. d–f Quantification of the amount of MDR1 or Survivin, normalized to the amount of β-actin. The results are representative of five independent experiments. *P < 0.01 compared to RPMI8226 cells (ANOVA with Dunnett’s test)
Further, to assess the role of MDR1 and Survivin in the melphalan resistance of RPMI8226/L-PAM cells, these cells were treated with MDR1 and Survivin siRNAs. Treatment of cells with MDR1 siRNA at a concentration sufficient to completely suppress MDR1 expression did not overcome their melphalan resistance (Fig. 4a, d, g, j). However, siRNA-mediated silencing of Survivin was found to partially overcome melphalan resistance in RPMI8226/L-PAM cells (Fig. 4b, e, h, k). In addition, RPMI8226 cells treated with Bim siRNA acquired melphalan resistance (Fig. 4c, f, i, m). These results suggest that the mechanism of drug resistance in RPMI8226/L-PAM cells may involve upregulation of Survivin expression and downregulation of Bim expression, but not upregulation of MDR1 expression. These results indicate that enhanced Survivin expression and decreased Bim expression may play a critical role in the mechanism of drug resistance in RPMI8226/L-PAM cells.
Effect of MDR1, Survivin, and Bim siRNAs on melphalan resistance. a, b RPMI8226/L-PAM cells treated with negative siRNA, MDR1 siRNA, or Survivin siRNA. c RPMI8226 cells treated with negative siRNA or Bim siRNA. a–c Total RNA was extracted, and the MDR1, Survivin, or Bim mRNA levels were determined by real-time PCR. The results are expressed as the test:control ratio after correction of the GAPDH mRNA levels. The results are representative of five independent experiments. *P < 0.01 vs. control (ANOVA with Dunnett’s test). d–f Whole-cell lysates were extracted and immunoblotted with antibodies against d MDR1, e Survivin, f Bim, and β-actin. g–i Quantification of the amount of g MDR1, h Survivin, or i Bim, normalized to the amounts of the β-actin. The results are representative of 5 independent experiments. *P < 0.01, compared to controls (ANOVA with Dunnett’s test). j, k RPMI8226/L-PAM cells were subsequently exposed to the indicated concentrations of melphalan, MDR1 siRNA, or Survivin siRNA. After incubation for 72 h, the number of dead cells was measured by trypan blue staining. The results are representative of five independent experiments. *P < 0.01 vs. untreated cells (analysis of variance with Dunnett’s test). l RPMI8226 cells were subsequently exposed to the indicated concentrations of melphalan or Bim siRNA. After incubation for 72 h, the number of dead cells was measured by trypan blue staining. The results are representative of five independent experiments. *P < 0.01 vs. untreated cells, #P < 0.01 vs. melphalan-treated cells (analysis of variance with Dunnett’s test)
Overexpression of HIF-1α via activation of ERK1/2, Akt, and NF-κB regulates Survivin and Bim expression in melphalan-resistant cells
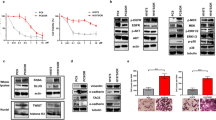
HIF-1α is involved in the transcription of various genes, such as Survivin and MDR1, and regulates apoptosis-inducing factors, such as Bim [34,35,36]. So far, the findings in this study indicate that melphalan resistance is achieved by downregulation of Bim and upregulation of Survivin, and activation of ERK1/2, Akt, and NF-κB. Further, to assess the expression of Survivin and Bim in melphalan-resistant RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells, we investigated the expression levels of HIF-1α. HIF-1α protein level and activity were increased in RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells compared to parent cells (Fig. 5a, b, and Supplementary Fig. S9a). We also examined whether inhibition of HIF-1α by echinomycin, an HIF-1α inhibitor, suppressed Survivin expression and increased Bim expression. Treatment with echinomycin inhibited Survivin expression and increased Bim expression in RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells, and combined treatment with melphalan and echinomycin overcame melphalan resistance (Fig. 5c–e and Supplementary Fig. S9b-c). In addition, connation of melphalan and HIF-1α siRNA induced cell death by suppressing Survivin expression and enhancing Bim expression in RPMI8226/L-PAM cells (Fig. 5f, g).
Melphalan-resistant cells overexpress HIF-1α and echinomycin overcomes melphalan resistance. a The cytoplasmic and nuclear fractions were extracted and then subjected to SDS-PAGE/immunoblotting with anti-HIF-1α antibody. Anti-β-actin and anti-lamin antibodies were used as internal standards. b RPMI8226/L-PAM cells constitutively activate HIF-1α. The nuclear fractions were extracted and analyzed for HIF-1α activity using HIF-1α transcription factor assay kit. The results are representative of five independent experiments. *P < 0.01 vs. RPMI8226 cells (ANOVA with Dunnett’s test). c Echinomycin inhibited HIF-1α activity. Cells were treated with 0.01, 0.1, or 1 nM echinomycin. After incubation for 24 h, the nuclear fractions were extracted and analyzed for HIF-1α activity using HIF-1α transcription factor assay kit. The results are representative of five independent experiments. *P < 0.01 vs. RPMI8226/L-PAM untreated cells (ANOVA with Dunnett’s test). d RPMI8226/L-PAM and ARH-77/L-PAM cells were exposed to the indicated concentrations of echinomycin. After incubation with inhibitors for 72 h, the cell lysates were extracted and then subjected to SDS-PAGE/immunoblotting with antibodies against Survivin, Bim, and β-actin. Anti-β-actin antibody was used as an internal standard. e RPMI8226/L-PAM and ARH-77/L-PAM cells were exposed to the indicated concentrations of melphalan or ecinomycin. After incubation for 72 h, the number of dead cells was counted by trypan blue staining. The results are representative of five independent experiments. *P < 0.01 vs. RPMI8226/L-PAM or ARH-77/L-PAM untreated cells (ANOVA with Dunnett’s test). f RPMI8226/L-PAM cells were treated with negative siRNA or HIF-1α siRNA. The cytoplasmic and nuclear fractions were extracted and then subjected to SDS-PAGE/immunoblotting with anti-HIF-1α, anti-Survivin, and anti-Bim antibodies. Anti-β-actin and anti-lamin antibodies were used as internal standards. g Cells were subsequently exposed to the indicated concentrations of melphalan, HIF-1α siRNA, or negative siRNA. After incubation for 72 h, the number of dead cells was measured by trypan blue staining. The results are representative of five independent experiments. *P < 0.01 vs. untreated cells (ANOVA with Dunnett’s test)
Next, we investigated whether inhibition of ERK1/2, Akt, or NF-κB suppresses HIF-1α expression. Treatment with U0126, LY294002, or DMF alone did not suppress the expression of HIF-1α (Fig. 6a, d). Double-inhibitor combinations moderately reduced the expression of HIF-1α, and triple-inhibitor combinations decreased HIF-1α expression to levels similar to those of parent cells (Fig. 6b, c, e, f).
U0126, LY294002, and DMF suppress HIF-1α expression in RPMI8226/L-PAM cells. a–c RPMI8226/L-PAM cells were exposed to the indicated concentrations of U0126, LY294002, or dimethyl fumarate (DMF). After incubation with inhibitors for 72 h, the cell lysates were extracted and then subjected to SDS-PAGE/immunoblotting with antibodies against HIF-1α and lamin. Anti-lamin antibody was used as an internal standard. d–f Quantification of the amount of HIF-1α, normalized to the amount of β-actin. The results are representative of five independent experiments. *P < 0.01 compared to RPMI8226 cells (ANOVA with Dunnett’s test)
Overexpression of HIF-1α correlates with poor prognosis in MM patients
We analyzed the relationship between expression levels of HIF-1α and prognosis of human MM patients using the SurvExpress database. We observed that the expression level of HIF-1α correlates with poor overall survival (Fig. 7), and thus concluded that HIF-1α overexpression is associated with poor prognosis and reduced drug sensitivity in MM patients.
Discussion
In this study, we demonstrate that melphalan resistance in RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells is associated with ERK1/2, Akt, and NF-κB activation, and inhibition of these signaling factors overcame melphalan resistance. Activation of ERK1/2 and Akt has been shown to suppress cyclophosphamide-induced cell death, and these specific inhibitors enhance sensitivity to cyclophosphamide [37]. In addition, perifosine, an Akt inhibitor, enhances the cytotoxicity of dexamethasone, melphalan, or doxorubicin in MM cell lines [38]. Alkylating agent-induced NF-κB activation is associated with chemoresistance, and suppression of NF-κB reverses the sensitivity to alkylating agents [39, 40]. Together, our previous study and this one show that, in melphalan-resistant cells, phosphorylation of c-Src induces ERK1/2, Akt, and NF-κB activation, and inhibition of c-Src by dasatinib resensitizes the cells to melphalan [31]. It was reported that macrophage inflammatory protein 1α (MIP-1α) or tumor necrosis factor α (TNFα) promote c-Src activation via binding to specific receptors in cancer cells [41]. In addition, activation of ERK1/2, Akt, and/or NF-κB by MIP-1α and/or TNFα autocrine loop is associated with melphalan resistance in MM cells [40, 42]. These findings indicated that activation of ERK1/2, Akt, and NF-κB contributes to melphalan resistance in MM cells.
Our previous study clearly indicated that overexpression of MDR1 and Survivin and decreased Bim expression are associated with melphalan resistance in RPMI8226/L-PAM cells. In the present study, we found that inhibition of ERK1/2, Akt, or NF-κB by U0126, LY294002, or DMF, respectively, suppressed MDR1 expression, but did not affect Survivin and Bim expression in RPMI8226/L-PAM cells, and then did not reverse melphalan resistance. Double-inhibitor combinations significantly inhibited Survivin expression, partiality enhanced Bim expression, and moderately overcame melphalan resistance. Triple-inhibitor combination overcame melphalan resistance by suppressing Survivin expression and enhancing Bim expression in RPMI8226/L-PAM cells. These results indicate that activation of ERK1/2, Akt, or NF-κB promotes the MDR1 expression, but MDR1 does not contribute to melphalan resistance. In addition, siRNA-mediated silencing of Survivin partially overcame melphalan resistance; however, melphalan resistance was not overcome by siRNA-mediated silencing of MDR1. Furthermore, parent cells treated with Bim siRNA acquired melphalan resistance. It has been reported that combination with valspodar (an MDR1 inhibitor), vincristine, adriamycin, and dexamethasone did not improve overall survival and progression free survival in recurring or refractory patients with MM, as compared to vincristine, adriamycin, and dexamethasone combination therapy in phase III trial [43]. It has also been reported that overexpression of Survivin and downregulation of Bim are correlated with poor prognosis in patients with MM [44]. Relapse patients of MM overexpress Survivin, and treatment with Survivin siRNA has been shown to enhance the cytotoxic effect of adriamycin, dexamethasone, and melphalan [45]. Lack of pro-apoptotic Bim function reduces the response to chemotherapy and overall survival in acute lymphoblastic lymphoma [46]. These findings suggest that overexpression of Survivin and decreased Bim expression may play an important role in melphalan resistance.
HIF-1α is known to be a transcription factor, and is related with various cellular functions, such as cell proliferation, survival, and drug resistance [47]. We clearly showed the RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells overexpressed HIF-1α compared with parent cells, and echinomycin overcame melphalan resistance by suppressing Survivin expression and enhancing Bim expression in RPMI8226/L-PAM, ARH-77/L-PAM, and HS-Sultan/L-PAM cells. In addition, treatment with HIF-1α siRNA reversed the melphalan sensitivity in RPMI8226/L-PAM cells. Moreover, inhibition of ERK1/2, Akt, and NF-κB by treatment with U0126, LY294002, and DMF significantly suppressed the expression of HIF-1α in RPMI8226/L-PAM cells. It has been reported that binding of HIF-1α to the Survivin promoter increases transcription of the Survivin gene [48]. In addition, induction of HIF-1α downregulates Bim expression at a transcriptional level [49]. Moreover, activation of the PI3K/Akt and MEK/ERK pathways enhances hypoxia-independent HIF-1α expression [50]. It has also been indicated that activation of NF-κB signaling by phosphatase of regenerating liver-3 promotes HIF-1α expression under normoxia and hypoxia conditions in gastric cancer cells [51]. These findings suggested that enhanced HIF-1α expression via activation of ERK1/2, Akt, and NF-κB leads to upregulation of Survivin and downregulation of Bim expression.
In this study, we found that MM patients showing high expression levels of HIF-1α had significantly lower overall survival rates than those showing low HIF-1α expression levels. Evidence shows that overexpression of HIF-1α promotes disease progression and drug resistance, thereby conducing to poor prognosis in MM patients. It was reported that myeloma patients with t(4;14) translocations, a poorer clinical outcome, showed significantly higher expression of HIF-1α, and that high expression of HIF-1α and EP300 are correlated with poor prognosis [11, 52]. In addition, hypoxia conditions enhanced tumor initiation and induced resistance to bortezomib and carfilzomib in MM cells [53, 54]. It was also reported that downregulation of HIF-1α expression enhanced sensitivity to bortezomib and lenelidomide [55, 56]. Furthermore, we found that our established melphalan-resistant MM cells showed decreased sensitivity to bortezomib (data not shown). These finding suggests that high expression of HIF-1α contributes to resistance to conventional anticancer drugs, such as melphalan, proteasome inhibitors, and immunomodulatory drugs, and inhibition of HIF-1α resensitized the sensitivity to those drugs in MM cells, therefore potentially improving the overall survival and progression-free survival of MM patients.
Our results also indicate that melphalan resistant HS-Sultan cells, a Burkitt’s lymphoma cell line, overexpress HIF-1α via activation of c-Src, ERK1/2, Akt, and NF-κB, and show enhanced expression of Survivin and decreased expression of Bim. In Burkitt’s lymphoma patients, the melphalan-containing regimen of the Haemato Oncology Foundation for Adults in The Netherlands (HOVON), with 69% of complete response, performed as well as other regimens, such as the French Lymphome Malins B (LMB) regimen and the German Berlin-Frankfurt-Munster (BFM) regimen, but with refractory disease of 21% and relapse rate of 9% [57]. In addition, patients with low Bim expression presented lower complete remission rate (24%) and shorter overall survival, and Burkitt’s lymphoma patients with high Survivin expression responded poorly to chemotherapy [58, 59]. Taken together, these findings suggest that overexpression of HIF-1α induces Survivin expression and decreases Bim expression, mediating chemoresistance and contributing to poor prognosis in Burkitt’s lymphoma patients. Inhibition of HIF-1α may overcome chemoresistance in B cell lymphomas such as Burkitt’s lymphoma.
In conclusion, we suggest that overexpression of Survivin and decreased Bim expression play an important role in MDR in melphalan-resistant cells via overexpression of HIF-1α and activation of ERK1/2, Akt, and NF-κB. Thus, regulation of these signaling molecules by inhibitors may be a viable approach for increasing the susceptibility of quiescent MM cells to chemotherapy, thereby reducing the incidence of minimal residual disease following MM cell therapy. Our findings suggest that inhibitors of HIF-1α, ERK1/2, Akt, and NF-κB may be useful anti-MDR agents for the treatment of melphalan-resistant MM.
References
Tai YT, Podar K, Mitsiades N, et al. CD40 induces human multiple myeloma cell migration via phosphatidylinositol 3-kinase/AKT/NF-kappa B signaling. Blood. 2003;101:2762–9.
Tohnya TM, Figg WD. Immunomodulation of multiple myeloma. Cancer Biol Ther. 2004;3:1060–1.
Dimopoulos MA, Goldschmidt H, Niesvizky R, et al. Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): an interim overall survival analysis of an open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18:1327–37.
National Cancer Institute Surveillance, Epidemiology, and End Results Program. https://seer.cancer.gov/statfacts/html/mulmy.html. Accessed 12 May 2018.
Ludwig H, Milosavljevic D, Zojer N, et al. Immunoglobulin heavy/light chain ratios improve paraprotein detection and monitoring, identify residual disease and correlate with survival in multiple myeloma patients. Leukemia. 2013;27:213–9.
Tsubaki M, Satou T, Itoh T, et al. Overexpression of MDR1 and survivin, and decreased Bim expression mediate multidrug-resistance in multiple myeloma cells. Leuk Res. 2012;36:1315–22.
Jimenez-Zepeda VH, Duggan P, Neri P, et al. Bortezomib-containing regimens (BCR) for the treatment of non-transplant eligible multiple myeloma. Ann Hematol. 2017;96:431–9.
Mimura N, Hideshima T, Anderson KC. Novel therapeutic strategies for multiple myeloma. Exp Hematol. 2015;43:732–41.
Manasanch EE, Orlowski RZ. Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol. 2017;14:417–33.
Gullà A, Di Martino MT, Gallo Cantafio ME, et al. A 13 mer LNA-i-miR-221 inhibitor restores drug sensitivity in melphalan-refractory multiple myeloma cells. Clin Cancer Res. 2016;22:1222–33.
Moreau P, Attal M, Facon T. Frontline therapy of multiple myeloma. Blood. 2015;125:3076–84.
Mateos MV, Richardson PG, Dimopoulos MA, et al. Effect of cumulative bortezomib dose on survival in multiple myeloma patients receiving bortezomib-melphalan-prednisone in the phase III VISTA study. Am J Hematol. 2015;90:314–9.
Kim MK, Kim K, Min CK, et al. A prospective, open-label, multicenter, observational study to evaluate the efficacy and safety of bortezomib-melphalan-prednisone as initial treatment for autologous stem cell transplantation-ineligible patients with multiple myeloma. Oncotarget. 2017;8:37605–18.
Offidani M, Corvatta L, Liberati AM, et al. Updated results of a phase 2 study of panobinostat combined with melphalan, thalidomide and prednisone (MPT) in relapsed/refractory multiple myeloma. Leuk Lymphoma. 2018;59:1271–3.
Zweegman S, van der Holt B, Mellqvist UH, et al. Melphalan, prednisone, and lenalidomide versus melphalan, prednisone, and thalidomide in untreated multiple myeloma. Blood. 2016;127:1109–16.
Mateos MV, Dimopoulos MA, Cavo M, et al. Daratumumab plus bortezomib, melphalan, and prednisone for untreated myeloma. N Eng J Med. 2018;378:518–28.
Vangsted AJ, Helm-Petersen S, Cowland JB, et al. Drug response prediction in high-risk multiple myeloma. Gene. 2018;644:80–6.
Tsuruo T, Naito M, Tomida A, et al. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci. 2003;94:15–21.
Eckford PD, Sharom FJ. ABC efflux pump-based resistance to chemotherapy drugs. Chem Rev. 2009;109:2989–3011.
Delbridge AR, Grabow S, Strasser A, et al. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 2016;16:99–109.
Tsubaki M, Takeda T, Yoshizumi M, et al. RANK-RANKL interactions are involved in cell adhesion-mediated drug resistance in multiple myeloma cell lines. Tumour Biol. 2016;37:9099–110.
Pennati M, Folini M, Zaffaroni N. Targeting survivin in cancer therapy: fulfilled promises and open questions. Carcinogenesis. 2007;28:1133–9.
Kojima H, Iida M, Yaguchi Y, et al. Enhancement of Cisplatin sensitivity in squamous cell carcinoma of the head and neck transfected with a survivin antisense gene. Arch Otolaryngol Head Neck Surg. 2006;132:682–5.
Zhang M, Mukherjee N, Bermudez RS, et al. Adenovirus-mediated inhibition of survivin expression sensitizes human prostate cancer cells to paclitaxel in vitro and in vivo. Prostate. 2005;64:293–302.
Di Marzo L, Desantis V, Solimando AG, et al. Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget. 2016;7:60698–711.
Jones VS, Huang RY, Chen LP, et al. Cytokines in cancer drug resistance: cues to new therapeutic strategies. Biochim Biophys Acta. 2016;1865:255–65.
Bentires-Alj M, Barbu V, Fillet M, et al. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene. 2003;22:90–7.
Zhang M, Yang J, Li F. Transcriptional and post-transcriptional controls of survivin in cancer cells: novel approaches for cancer treatment. J Exp Clin Cancer Res. 2003;25:391–402.
Hayun M, Naor Y, Weil M, et al. The immunomodulator AS101 induces growth arrest and apoptosis in multiple myeloma: association with the Akt/survivin pathway. Biochem Pharmacol. 2006;72:1423–31.
Li F, Sethi G. Targeting transcription factor NF-kappaB to overcome chemoresistance and radioresistance in cancer therapy. Biochim Biophys Acta. 2010;1805:167–80.
Tsubaki M, Komai M, Itoh T, et al. By inhibiting Src, verapamil and dasatinib overcome multidrug resistance via increased expression of Bim and decreased expressions of MDR1 and survivin in human multidrug-resistant myeloma cells. Leuk Res. 2014;38:121–30.
Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250.
Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8.
Lanvin O, Monferran S, Delmas C, et al. Radiation-induced mitotic cell death and glioblastoma radioresistance: a new regulating pathway controlled by integrin-linked kinase, hypoxia-inducible factor 1 alpha and survivin in U87 cells. Eur J Cancer. 2013;49:2884–91.
Schöning JP, Monteiro M, Gu W. Drug resistance and cancer stem cells: the shared but distinct roles of hypoxia-inducible factors HIF1α and HIF2α. Clin Exp Pharmacol Physiol. 2017;44:153–61.
Whelan KA, Schwab LP, Karakashev SV, et al. The oncogene HER2/neu (ERBB2) requires the hypoxia-inducible factor HIF-1 for mammary tumor growth and anoikis resistance. J Biol Chem. 2013;288:15865–77.
Yang X, Liu Y, Liu J, et al. Cyclophosphamide-induced apoptosis in A431 cells is inhibited by fucosyltransferase IV. J Cell Biochem. 2011;112:1376–83.
Hideshima T, Catley L, Yasui H, et al. Perifosine, an oral bioactive novel alkylphospholipid, inhibits Akt and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Blood. 2006;107:4053–62.
Meinel FG, Mandl-Weber S, Baumann P, et al. The novel, proteasome-independent NF-kappaB inhibitor V1810 induces apoptosis and cell cycle arrest in multiple myeloma and overcomes NF-kappaB-mediated drug resistance. Mol Cancer Ther. 2010;9:300–10.
Tsubaki M, Komai M, Itoh T, et al. Inhibition of the tumour necrosis factor-alpha autocrine loop enhances the sensitivity of multiple myeloma cells to anticancer drugs. Eur J Cancer. 2013;49:3708–17.
Liu ST, Pham H, Pandol SJ, et al. Src as the link between inflammation and cancer. Front Physiol. 2014;4:416.
Tsubaki M, Takeda T, Tomonari Y, et al. The MIP-1α autocrine loop contributes to decreased sensitivity to anticancer drugs. J Cell Physiol. 2018;233:4258–71.
Friedenberg WR, Rue M, Blood EA, et al. Phase III study of PSC-833 (valspodar) in combination with vincristine, doxorubicin, and dexamethasone (valspodar/VAD) versus VAD alone in patients with recurring or refractory multiple myeloma (E1A95): a trial of the Eastern Cooperative Oncology Group. Cancer . 2006;106:830–8.
Romagnoli M, Séveno C, Wuillème-Toumi S, et al. The imbalance between Survivin and Bim mediates tumour growth and correlates with poor survival in patients with multiple myeloma. Br J Haematol. 2009;145:180–9.
Romagnoli M, Trichet V, David C, et al. Significant impact of survivin on myeloma cell growth. Leukemia. 2007;21:1070–8.
Gagné V, Rousseau J, Labuda M, et al. Bim polymorphisms: influence on function and response to treatment in children with acute lymphoblastic leukemia. Clin Cancer Res. 2013;19:5240–9.
Rohwer N, Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist Updates. 2011;14:191–201.
Chen YQ, Zhao CL, Li W. Effect of hypoxia-inducible factor-1alpha on transcription of survivin in non-small cell lung cancer. J Exp Clin Cancer Res. 2009;28:29.
Sionov RV, Vlahopoulos SA, Granot Z. Regulation of Bim in health and disease. Oncotarget. 2015;6:23058–134.
Wan J, Wu W. Hyperthermia induced HIF-1a expression of lung cancer through AKT and ERK signaling pathways. J Exp Clin Cancer Res. 2016;35:119.
Zhang C, Tian W, Meng L, et al. PRL-3 promotes gastric cancer migration and invasion through a NF-κB-HIF-1α-miR-210 axis. J Mol Med. 2016;94:401–15.
Viziteu E, Grandmougin C, Goldschmidt H, et al. Chetomin, targeting HIF-1α/p300 complex, exhibits antitumour activity in multiple myeloma. Br J Cancer. 2016;114:519–23.
Muz B, de la Puente P, Azab F, et al. Hypoxia promotes stem cell-like phenotype in multiple myeloma cells. Blood Cancer J. 2014;4:e262.
Kocemba-Pilarczyk KA, Ostrowska B, Trojan S, et al. Targeting the hypoxia pathway in malignant plasma cells by using 17-allylamino-17-demethoxygeldanamycin. Acta Biochim Pol. 2018;65:101–9.
Borsi E, Perrone G, Terragna C, et al. Hypoxia inducible factor-1 alpha as a therapeutic target in multiple myeloma. Oncotarget. 2014;5:1779–92.
Storti P, Toscani D, Airoldi I, et al. The anti-tumoral effect of lenalidomide is increased in vivo by hypoxia-inducible factor (HIF)-1α inhibition in myeloma cells. Haematologica. 2016;101:e107–10.
Oosten LEM, Chamuleau MED, Thielen FW, et al. Treatment of sporadic Burkitt lymphoma in adults, a retrospective comparison of four treatment regimens. Ann Hematol. 2018;97:255–66.
Richter-Larrea JA, Robles EF, Fresquet V, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–42.
Zyada MM. Relationship of survivin to clinical drug resistance in Burkitt’s lymphoma of the head and neck region. Med Oncol. 2011;28:1565–9.
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research (C)(Grant number 15K08116), Grant-in-Aid for Young Scientists (B)(Grant number 16K18965) from the Japan Society for the Promotion of Science (JSPS) and by Ministry of Education, Culture, Sports, Science, and Technology (MEXT)-Supported Program for the Strategic Research Foundation at Private Universities, 2014–2018 (Grant number S1411037).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tsubaki, M., Takeda, T., Tomonari, Y. et al. Overexpression of HIF-1α contributes to melphalan resistance in multiple myeloma cells by activation of ERK1/2, Akt, and NF-κB. Lab Invest 99, 72–84 (2019). https://doi.org/10.1038/s41374-018-0114-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0114-8
This article is cited by
-
Osteocyte-derived exosomes confer multiple myeloma resistance to chemotherapy through acquisition of cancer stem cell-like features
Leukemia (2023)
-
Targeting hypoxia in solid and haematological malignancies
Journal of Experimental & Clinical Cancer Research (2022)
-
Bim downregulation by activation of NF-κB p65, Akt, and ERK1/2 is associated with adriamycin and dexamethasone resistance in multiple myeloma cells
Clinical and Experimental Medicine (2022)
-
Pharmacodynamic mechanisms of anti-inflammatory drugs on the chemosensitization of multidrug-resistant cancers and the pharmacogenetics effectiveness
Inflammopharmacology (2021)
-
Crosstalk between endoplasmic reticulum stress and oxidative stress: a dynamic duo in multiple myeloma
Cellular and Molecular Life Sciences (2021)