Abstract
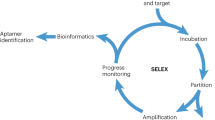
Aptamers are single-stranded oligonucleotides that are in vitro-selected to recognize their target molecule with high affinity and specificity. As they consist of the four canonical nucleobases, their chemical diversity is limited, which in turn limits the addressable target spectrum. Introducing chemical modifications into nucleic acid libraries increases the interaction capabilities of the DNA and thereby the target spectrum. Here, we describe a protocol to select nucleobase-modified aptamers by using click chemistry (CuAAC) to introduce the preferred chemical modification. The use of click chemistry to modify the DNA library enables the introduction of a wide range of possible functionalities, which can be customized to the requirements of the target molecule and the desired application. This protocol yields modified DNA aptamers with extended interaction properties that are not accessible with the canonical set of nucleotides. After synthesis of the starting library containing a commercially available, alkyne-modified uridine (5-ethynyl-deoxyuridine (EdU)) instead of thymidine, the library is functionalized with the modification of choice by CuAAC. The thus-modified DNA is incubated with the target molecule and the best binding sequences are recovered. The chemical modification is removed during the amplification process. Therefore, this protocol is compatible with conventional amplification procedures and avoids enzymatic incompatibility problems associated with more extensive nucleobase modifications. After single-strand generation, the modification is reintroduced into the enriched library, which can then be subjected to the subsequent selection cycle. The duration of each selection cycle as outlined in the protocol is ∼1 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Gierlich, J., Burley, G.A., Gramlich, P.M.E., Hammond, D.M. & Carell, T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 8, 3639–3642 (2006).
El-Sagheer, A.H. & Brown, T. Click chemistry with DNA. Chem. Soc. Rev. 39, 1388 (2010).
Nikic, I., Kang, J.H., Girona, G.E., Aramburu, I.V. & Lemke, E.A. Labeling proteins on live mammalian cells using click chemistry. Nat. Protoc. 10, 780–791 (2015).
Sirbu, B.M., Couch, F.B. & Cortez, D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7, 594–605 (2012).
Kukwikila, M., Gale, N., El-Sagheer, A.H., Brown, T. & Tavassoli, A. Assembly of a biocompatible triazole-linked gene by one-pot click-DNA ligation. Nat. Chem. 9, 1089–1098 (2017).
Gierlich, J. et al. Synthesis of highly modified DNA by a combination of PCR with alkyne-bearing triphosphates and click chemistry. Chemistry 13, 9486–9494 (2007).
Tolle, F., Brändle, G.M., Matzner, D. & Mayer, G. A versatile approach towards nucleobase-modified aptamers. Angew. Chem. 54, 10971–10974 (2015).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A.D. & Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Taylor, A.I. & Holliger, P. Directed evolution of artificial enzymes (XNAzymes) from diverse repertoires of synthetic genetic polymers. Nat. Protoc. 10, 1625–1642 (2015).
Mayer, G. et al. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 5, 1993–2004 (2010).
Sefah, K., Shangguan, D., Xiong, X., O'Donoghue, M.B. & Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 5, 1169–1185 (2010).
Pfeiffer, F. & Mayer, G. Selection and biosensor application of aptamers for small molecules. Front. Chem. 4, 25 (2016).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5, e15004 (2010).
Davies, D.R. et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA 109, 19971–19976 (2012).
Kimoto, M., Yamashige, R., Matsunaga, K., Yokoyama, S. & Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 31, 453–457 (2013).
Pfeiffer, F., Rosenthal, M., Siegl, J., Ewers, J. & Mayer, G. Customised nucleic acid libraries for enhanced aptamer selection and performance. Curr. Opin. Biotechnol. 48, 111–118 (2017).
Sefah, K. et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 111, 1449–1454 (2014).
Mendonsa, S.D. & Bowser, M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 126, 20–21 (2004).
Daniels, D.A., Chen, H., Hicke, B.J., Swiderek, K.M. & Gold, L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 100, 15416–15421 (2003).
Gawande, B.N. et al. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. U.S.A. 114, 2898–2903 (2017).
Hollenstein, M. DNA catalysis: the chemical repertoire of DNAzymes. Molecules 20, 20777–20804 (2015).
Zhou, C. et al. DNA-catalyzed amide hydrolysis. J. Am. Chem. Soc. 138, 2106–2109 (2016).
Lipi, F., Chen, S., Chakravarthy, M., Rakesh, S. & Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: pros & cons, and comprehensive selection strategies. RNA Biol. 13, 1232–1245 (2016).
Matsunaga, K.I., Kimoto, M. & Hirao, I. High-affinity DNA aptamer generation targeting von Willebrand factor A1-domain by genetic alphabet expansion SELEX (ExSELEX) using two types of libraries composed of five different bases. J. Am. Chem. Soc. 139, 324–334 (2017).
Kimoto, M., Matsunaga, K. & Hirao, I. DNA aptamer generation by genetic alphabet expansion SELEX (ExSELEX) using an unnatural base pair system. Methods Mol. Biol. 1380, 47–60 (2016).
Latham, J.A., Johnson, R. & Toole, J.J. The application of a modified nucleotide in aptamer selection: novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res. 22, 2817–2822 (1994).
Tolle, F., Wilke, J., Wengel, J. & Mayer, G. By-product formation in repetitive PCR amplification of DNA libraries during SELEX. PLoS ONE 9, e114693 (2014).
Caruthers, M.H. Chemical synthesis of DNA and DNA analogs. Acc. Chem. Res. 24, 278–284 (1991).
Hall, L.M., Gerowska, M. & Brown, T. A highly fluorescent DNA toolkit: synthesis and properties of oligonucleotides containing new Cy3, Cy5 and Cy3B monomers. Nucleic Acids Res. 40, e108 (2012).
Suzuki, T. et al. Rapid discovery of highly potent and selective inhibitors of histone deacetylase 8 using click chemistry to generate candidate libraries. J. Med. Chem. 55, 9562–9575 (2012).
Shim, J.-Y., Bertalovitz, A.C. & Kendall, D.A. Identification of essential cannabinoid-binding domains: structural insights into early dynamic events in receptor activation. J. Biol. Chem. 286, 33422–33435 (2011).
Hua, T. et al. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 547, 468–471 (2017).
Hua, T. et al. Crystal structure of the human cannabinoid receptor CB1. Cell 167, 750–762 (2016).
Shao, Z. et al. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature 540, 602–606 (2016).
Hipolito, C.J., Hollenstein, M., Lam, C.H. & Perrin, D.M. Protein-inspired modified DNAzymes: dramatic effects of shortening side-chain length of 8-imidazolyl modified deoxyadenosines in selecting RNaseA mimicking DNAzymes. Org. Biomol. Chem. 9, 2266–2273 (2011).
Besanceney-Webler, C. et al. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew. Chem. Int. Ed. Engl. 50, 8051–8056 (2011).
Stoltenburg, R., Nikolaus, N. & Strehlitz, B. Capture-SELEX: selection of DNA aptamers for aminoglycoside antibiotics. J. Analyt. Methods Chem. 2012, 415697 (2012).
Nutiu, R. & Li, Y. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. Engl. 44, 1061–1065 (2005).
Yang, K.A., Pei, R., Stefanovic, D. & Stojanovic, M.N. Optimizing cross-reactivity with evolutionary search for sensors. J. Am. Chem. Soc. 134, 1642–1647 (2012).
Hermanson, G.T. Bioconjugate techniques (Academic press, 2013).
Srisawat, C. & Engelke, D.R. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA 7, 632–641 (2001).
Bittker, J.A., Le, B.V. & Liu, D.R. Nucleic acid evolution and minimization by nonhomologous random recombination. Nat. Biotechnol. 20, 1024–1029 (2002).
Tahiri-Alaoui, A. et al. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucleic Acids Res. 30, e45 (2002).
Stoltenburg, R., Reinemann, C. & Strehlitz, B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 383, 83–91 (2005).
Wang, C., Yang, G., Luo, Z. & Ding, H. In vitro selection of high-affinity DNA aptamers for streptavidin. Acta Biochim. Biophys. Sin. 41, 335–340 (2009).
Jenison, R.D., Gill, S.C., Pardi, A. & Polisky, B. High-resolution molecular discrimination by RNA. Science 263, 1425–1429 (1994).
Hamedani, N.S. et al. Capture and release (CaR): a simplified procedure for one-tube isolation and concentration of single-stranded DNA during SELEX. Chem. Commun. 51, 1135–1138 (2015).
Avci-Adali, M., Paul, A., Wilhelm, N., Ziemer, G. & Wendel, H.P. Upgrading SELEX technology by using lambda exonuclease digestion for single-stranded DNA generation. Molecules 15, 1–11 (2009).
McKeague, M. et al. Comprehensive analytical comparison of strategies used for small molecule aptamer evaluation. Anal. Chem. 87, 8608–8612 (2015).
Jauset Rubio, M. et al. beta-Conglutin dual aptamers binding distinct aptatopes. Anal. Bioanal. Chem. 408, 875–884 (2015).
Kellenberger, C.A. et al. A minimalist biosensor: quantitation of cyclic di-GMP using the conformational change of a riboswitch aptamer. RNA Biol. 12, 1189–1197 (2015).
Tolle, F. & Mayer, G. Preparation of SELEX samples for next-generation sequencing. Methods Mol. Biol. 1380, 77–84 (2016).
Blank, M. Next-generation analysis of deep sequencing data: bringing light into the black box of SELEX experiments. Methods Mol. Biol. 1380, 85–95 (2016).
Thiel, W.H. & Giangrande, P.H. Analyzing HT-SELEX data with the Galaxy Project tools—a web based bioinformatics platform for biomedical research. Methods 97, 3–10 (2016).
Beier, R., Boschke, E. & Labudde, D. New strategies for evaluation and analysis of SELEX experiments. BioMed Res. Intl. 2014, 849743 (2014).
Nguyen Quang, N., Perret, G. & Duconge, F. Applications of high-throughput sequencing for in vitro selection and characterization of aptamers. Pharmaceuticals 9 http://dx.doi.org/10.3390/ph9040076 (2016).
Phosphate buffer. Cold Spring Harb Protoc http://dx.doi.org/10.1101/pdb.rec8543 (2006).
Ingale, S.A., Mei, H., Leonard, P. & Seela, F. Ethynyl side chain hydration during synthesis and workup of 'clickable' oligonucleotides: bypassing acetyl group formation by triisopropylsilyl protection. J. Org. Chem. 78, 11271–11282 (2013).
Tolle, F., Rosenthal, M., Pfeiffer, F. & Mayer, G. Click reaction on solid phase enables high fidelity synthesis of nucleobase-modified DNA. Bioconjug. Chem. 27, 500–503 (2016).
Goujon, M. et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38, W695–W699 (2010).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Bailey, T.L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Acknowledgements
This work was made possible through funding by BMWi-ZIM (grant no. KF3058901SK2) and the Deutsche Forschungsgemeinschaft (grant nos. MA3442/4-1 and MA3442/4-2). We thank J.L. Schultze, K. Händler and M. Beyer (Life and Medical Sciences Institute and German Center for Neurodegenerative Diseases, Bonn) for their support regarding next-generation sequencing and M. Blank and C. Gröber (AptaIT, Munich, Germany) for their support regarding the NGS analysis. We also thank Protzek Gesellschaft für biomedizinische Technik mbH (Lörrach, Germany) for support and helpful discussions.
Author information
Authors and Affiliations
Contributions
F.P., F.T. and G.M. conceived and designed the protocol and wrote the manuscript. F.P., F.T. and M.R. performed the experiments. G.B. and J.E. synthesized the azides used in this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Pfeiffer, F., Tolle, F., Rosenthal, M. et al. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat Protoc 13, 1153–1180 (2018). https://doi.org/10.1038/nprot.2018.023
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.023
This article is cited by
-
Tryptophan-like side chain holding aptamers inhibit respiratory syncytial virus infection of lung epithelial cells
Scientific Reports (2023)
-
In vitro selection of aptamers and their applications
Nature Reviews Methods Primers (2023)
-
The selection of a hydrophobic 7-phenylbutyl-7-deazaadenine-modified DNA aptamer with high binding affinity for the Heat Shock Protein 70
Communications Chemistry (2023)
-
A high-dimensional microfluidic approach for selection of aptamers with programmable binding affinities
Nature Chemistry (2023)
-
Aptamers as Recognition Elements for Electrochemical Detection of Exosomes
Chemical Research in Chinese Universities (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.