Abstract
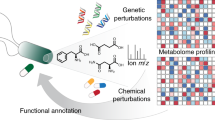
Cell-based high-content screens are increasingly used to discover bioactive small molecules. However, identifying the mechanism of action of the selected compounds is a major bottleneck. Here we describe a protocol consisting of experimental and computational steps to identify the cellular pathways modulated by chemicals, and their mechanism of action. The multiparametric profiles from a 'query' chemical screen are used as constraints to select genes with similar profiles from a 'reference' genetic screen. In our case, the query screen is the intracellular survival of mycobacteria and the reference is a genome-wide RNAi screen of endocytosis. The two disparate screens are bridged by an 'intermediate' chemical screen of endocytosis, so that the similarity in the multiparametric profiles between the chemical and the genetic perturbations can generate a testable hypothesis of the cellular pathways modulated by the chemicals. This approach is not assay specific, but it can be broadly applied to various quantitative, multiparametric data sets. Generation of the query system takes 3–6 weeks, and data analysis and integration with the reference data set takes an 3 additional weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Collinet, C. et al. Systems survey of endocytosis by multiparametric image analysis. Nature 464, 243–249 (2010).
Danuser, G. Computer vision in cell biology. Cell 147, 973–978 (2011).
Stockwell, B.R. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 1, 116–125 (2000).
Bickle, M. The beautiful cell: high-content screening in drug discovery. Anal. Bioanal. Chem. 398, 219–226 (2010).
Young, D.W. et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat. Chem. Biol. 4, 59–68 (2008).
Swinney, D.C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011).
Merino, A., Bronowska, A.K., Jackson, D.B. & Cahill, D.J. Drug profiling: knowing where it hits. Drug Discov. Today 15, 749–756 (2010).
Cong, F., Cheung, A.K. & Huang, S.M. Chemical genetics-based target identification in drug discovery. Annu. Rev. Pharmacol. Toxicol. 52, 57–78 (2012).
Stockwell, B.R. Exploring biology with small organic molecules. Nature 432, 846–854 (2004).
Schenone, M., Dancik, V., Wagner, B.K. & Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 9, 232–240 (2013).
Sundaramurthy, V. et al. Integration of chemical and RNAi multiparametric profiles identifies triggers of intracellular mycobacterial killing. Cell Host Microbe 13, 129–142 (2013).
Kittler, R. et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9, 1401–1412 (2007).
Neumann, B. et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 (2010).
Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78–86 (2005).
Paul, P. et al. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell 145, 268–283 (2011).
Yin, Z. et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat. Cell Biol. 15, 860–871 (2013).
WHO. Global Tuberculosis Report. (World Health Organization, 2013).
Zumla, A., Nahid, P. & Cole, S.T. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 12, 388–404 (2013).
Goodwin, P.J., Meyerhardt, J.A. & Hursting, S.D. Host factors and cancer outcome. J. Clin. Oncol. 28, 4019–4021 (2010).
Gutierrez, M.G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Kumar, D. et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140, 731–743 (2010).
Armstrong, J.A. & Hart, P.D. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 134, 713–740 (1971).
Russell, D.G. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2, 569–577 (2001).
Jayachandran, R. et al. Survival of mycobacteria in macrophages is mediated by coronin 1-dependent activation of calcineurin. Cell 130, 37–50 (2007).
Koul, A., Herget, T., Klebl, B. & Ullrich, A. Interplay between mycobacteria and host signalling pathways. Nat. Rev. Microbiol. 2, 189–202 (2004).
Behar, S.M., Divangahi, M. & Remold, H.G. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 8, 668–674 (2010).
Chen, M., Gan, H. & Remold, H.G. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J. Immunol. 176, 3707–3716 (2006).
Keane, J., Remold, H.G. & Kornfeld, H. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164, 2016–2020 (2000).
Agaisse, H. et al. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309, 1248–1251 (2005).
Philips, J.A., Rubin, E.J. & Perrimon, N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309, 1251–1253 (2005).
Dhandayuthapani, S. et al. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol. Microbiol. 17, 901–912 (1995).
Carpenter, A.E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Kamentsky, L. et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011).
Berthold, MR et al. KNIME: The Konstanz Information Miner (Springer, 2007).
Stoter, M. et al. CellProfiler and KNIME: open source tools for high content screening. Methods Mol. Biol. 986, 105–122 (2013).
Kittler, R. et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods 4, 337–344 (2007).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank J. Pieters (Biozentrum, Basel) for M. bovis BCG-GFP strain and A. Roesen-Wolff (Technische Universität Dresden (TUD)) for the help with monocyte isolation. We are indebted to High-Throughput Technology Development Studio (HT-TDS), MPI-CBG for expert technical assistance, to the High-Performance Computing Center at TUD and the Computer Department for outstanding IT support and to H. Nonaka for critical comments on the manuscript. The work was supported by the EU-FP7–funded projects NATT, PHAGOSYS and APO-SYS, and by the Max Planck Society.
Author information
Authors and Affiliations
Contributions
V.S. and M.Z. conceived the project; V.S., R.B., M.S. and N.T. developed and performed the experimental steps; M.C. and Y.K. developed the computational tools and performed the analysis; and M.B. and M.Z. coordinated and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Organization of the endosomics database.
The endosomics database (http://endosomics.mpi-cbg.de) is a repository for the high content data from the genome-wide screen of endocytosis. Quantitative multi-parametric profile for individual genes can be searched and exported. This figure details the central aspects of the organization and representation of the dataset.
Supplementary Figure 2 Screenshots from http://endosomics.mpi-cbg.de/gws/search (version 0.7.8.3).
This tool is used to compare the multi-parametric profiles from the query or intermediate systems with the endocytosis genome-wide reference system. (a) Users can generate the multi-parametric profiles for the query or intermediate system through the interactive graphic window. (b) Screen-shot of the list of parameters used in the endocytosis reference dataset. (c) Screenshot of the window to feed in correlation and phenoscore values.
Supplementary Figure 3 Screenshots from MotionTracking software (version 8.81.15) for the comparison of multiparametric profiles from reference dataset provided by the users.
(a) Screenshot of the step involved in importing an external reference dataset. (b-d) Screenshots of the steps involved in computing the phenoscore and correlations.
Supplementary information
Supplementary Figure 1
Organization of the endosomics database. (PDF 2416 kb)
Supplementary Figure 2
Screenshots from http://endosomics.mpi-cbg.de/gws/search (version 0.7.8.3). (PDF 1669 kb)
Supplementary Figure 3
Screenshots from MotionTracking software (version 8.81.15) for the comparison of multiparametric profiles from reference dataset provided by the users. (PDF 731 kb)
Supplementary Discussion
(PDF 1139 kb)
Supplementary Data 1
This is a compressed file that contains a sample MotionTracking project, nature_protocols.mtj, and some test images of mycobacteria infected human primary macrophages. Users are required to open the .mtj file in MotionTracking to access the images. Mycobacteria and cells have been pre-identified and the object search parameters are stored in the file nature_protocols_mt.srp. The parameters for object statistics are provided in nature_protocols_mt.stp. (ZIP 25508 kb)
Supplementary Data 2
This is a compressed file that contains a sample CellProfiler project, TbExample_pipeline.cp and some test images of mycobacteria-infected human primary macrophages that have been stained for lysosomes. (ZIP 16469 kb)
Supplementary Data 3
This is a compressed file that contains a .csv file having normalized multiparametric profiles of a subset of 1000 genes from the genome-wide endocytosis screen1. (ZIP 158 kb)
Rights and permissions
About this article
Cite this article
Sundaramurthy, V., Barsacchi, R., Chernykh, M. et al. Deducing the mechanism of action of compounds identified in phenotypic screens by integrating their multiparametric profiles with a reference genetic screen. Nat Protoc 9, 474–490 (2014). https://doi.org/10.1038/nprot.2014.027
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.027
This article is cited by
-
Image-based profiling for drug discovery: due for a machine-learning upgrade?
Nature Reviews Drug Discovery (2021)
-
A semi-automated, KNIME-based workflow for biofilm assays
BMC Microbiology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.