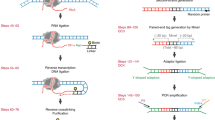
Abstract
Eviction or destabilization of nucleosomes from chromatin is a hallmark of functional regulatory elements in eukaryotic genomes. Historically identified by nuclease hypersensitivity, these regulatory elements are typically bound by transcription factors or other regulatory proteins. FAIRE (formaldehyde-assisted isolation of regulatory elements) is an alternative approach to identify these genomic regions and has proven successful in a multitude of eukaryotic cell and tissue types. Cells or dissociated tissues are cross-linked briefly with formaldehyde, lysed and sonicated. Sheared chromatin is subjected to phenol/chloroform extraction and the isolated DNA, typically encompassing 1–3% of the human genome, is purified. We provide guidelines for quantitative analysis by PCR, microarrays or next-generation sequencing. Regulatory elements enriched by FAIRE have high concordance with those identified by nuclease hypersensitivity or chromatin immunoprecipitation (ChIP), and the entire procedure can be completed in 3 d. FAIRE has low technical variability, which allows its usage in large-scale studies of chromatin from normal or diseased tissues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
13 January 2014
The authors have added some new information regarding the types of species and cell types in which formaldehyde-assisted isolation of regulatory elements (FAIRE) has been successfully performed, according to recently published reports. This information has been appended to the PDF version of the article.
References
Boyle, A.P. et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 21, 456–464 (2011).
Crawford, G.E. et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16, 123–131 (2006).
Boyle, A.P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Song, L. & Crawford, G.E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010 doi:10.1101/pdb.prot5384 (2010).
Keene, M.A., Corces, V., Lowenhaupt, K. & Elgin, S.C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proc. Natl. Acad. Sci. USA 78, 143–146 (1981).
McGhee, J.D., Wood, W.I., Dolan, M., Engel, J.D. & Felsenfeld, G. A 200 base pair region at the 5′ end of the chicken adult β-globin gene is accessible to nuclease digestion. Cell 27 (1, Part 2), 45–55 (1981).
Felsenfeld, G. & Groudine, M. Controlling the double helix. Nature 421, 448–453 (2003).
Gross, D.S. & Garrard, W.T. Nuclease hypersensitive sites in chromatin. Ann. Rev. Biochem. 57, 159–197 (1988).
Stalder, J. et al. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20, 451–460 (1980).
Hogan, G.J., Lee, C.-K. & Lieb, J.D. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2, e158 (2006).
Giresi, P.G., Kim, J., McDaniell, R.M., Iyer, V.R. & Lieb, J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007).
Giresi, P.G. & Lieb, J.D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods 48, 233–239 (2009).
Gaulton, K.J. et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 (2010).
Nagy, P.L., Cleary, M.L., Brown, P.O. &, Lieb J.D. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl. Acad. Sci. USA 100, 6364–6369 (2003).
Song, L. et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21, 1757–1767 (2011).
Ponts, N. et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20, 228–238 (2010).
Louwers, M. et al. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 21, 832–842 (2009).
Langmead, B., Trapnell, C., Pop, M. &, Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Rashid, N., Giresi, P.G., Ibrahim, J.G. & Lieb J.D. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 12, R67 (2011).
ENCODE Project Consortium. et al. A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046 (2011).
Birney, E., Lieb, J.D., Furey, T.S., Crawford, G.E. & Iyer, V.R. Allele-specific and heritable chromatin signatures in humans. Hum. Mol. Genet. 19, R204 (2010).
Hurtado, A., Holmes, K.A., Ross-Innes, C.S., Schmidt, D. & Carroll, J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 (2011).
Eeckhoute, J. et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 19, 372–380 (2009).
Egelhofer, T.A. et al. An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 18, 91–93 (2011).
Li, Q, Brown, J.B., Huang, H. & Bickel, P. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Lassmann, T., Hayashizaki, Y. & Daub, C.O. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics 25, 2839–2840 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows ∀ Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Boyle, A.P., Guinney, J., Crawford, G.E. & Furey, T.S. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24, 2537–2538 (2008).
Haring, M. et al. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3, 11 (2007).
Lee, T.I., Johnstone, S.E. & Young, R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006).
Ren, B. & Dynlacht, B.D. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 376, 304–315 (2004).
Oberley, M.J., Tsao, J., Yau, P. & Farnham, P.J. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376, 315–334 (2004).
Oberley, M.J. & Farnham, P.J. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 371, 577–596 (2003).
Lieb, J.D. Genome-wide mapping of protein-DNA interactions by chromatin immunoprecipitation and DNA microarray hybridization. Methods Mol. Biol. 224, 99–109 (2003).
Ciccone, D.N., Morshead, K.B. & Oettinger, M.A. Chromatin immunoprecipitation in the analysis of large chromatin domains across murine antigen receptor loci. Methods Enzymol. 376, 334–348 (2004).
Chaya, D. & Zaret, K.S. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol. 376, 361–372 (2004).
Buck, M.J. & Lieb, J.D. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83, 349–360 (2004).
Bernstein, B.E., Humphrey, E.L., Liu, C.L. & Schreiber, S.L. The use of chromatin immunoprecipitation assays in genome-wide analyses of histone modifications. Methods Enzymol. 376, 349–360 (2004).
Bannister, A.J. & Kouzarides, T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 376, 269–288 (2004).
Nammo, T., Rodriguez-Segui, S.A. & Ferrer, J. Mapping open chromatin with formaldehyde-assisted isolation of regulatory elements. Methods Mol. Biol. 791 (1940-6029 (Electronic)), 287–296 (2011).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Buck, M.J., Nobel, A.B. & Lieb, J.D. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 6, R97 (2005).
Sun, W., Buck, M., Patel, M. & Davis, I.J. Improved ChIP-chip analysis by a mixture model approach. BMC Bioinformatics 10, 173 (2009).
Schmittgen, T.D. & Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Fujita, P.A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39 (Database issue): D876–D882 (2011).
Acknowledgements
We acknowledge members of the Lieb and Davis labs for their constructive feedback. Support for this work was provided by ENCODE grant U54HG004563 from the National Human Genome Research Institute.
Author information
Authors and Affiliations
Contributions
The work presented here was carried out in collaboration between all authors. P.G.G. and J.M.S. designed and improved the method. J.M.S., P.G.G., I.J.D. and J.D.L. wrote the manuscript. All authors have contributed to, reviewed and approved of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Simon, J., Giresi, P., Davis, I. et al. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7, 256–267 (2012). https://doi.org/10.1038/nprot.2011.444
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.444
This article is cited by
-
Glucocorticoid receptor-NECAB1 axis can negatively regulate insulin secretion in pancreatic β-cells
Scientific Reports (2023)
-
Are extraordinary nucleosome structures more ordinary than we thought?
Chromosoma (2023)
-
Diurnal RNAPII-tethered chromatin interactions are associated with rhythmic gene expression in rice
Genome Biology (2022)
-
CENP-N promotes the compaction of centromeric chromatin
Nature Structural & Molecular Biology (2022)
-
Optimized assay for transposase-accessible chromatin by sequencing (ATAC-seq) library preparation from adult Drosophila melanogaster neurons
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.