Abstract
The effect of alterations in intestinal microbiota on microbial metabolites and on disease processes such as graft-versus-host disease (GVHD) is not known. Here we carried out an unbiased analysis to identify previously unidentified alterations in gastrointestinal microbiota–derived short-chain fatty acids (SCFAs) after allogeneic bone marrow transplant (allo-BMT). Alterations in the amount of only one SCFA, butyrate, were observed only in the intestinal tissue. The reduced butyrate in CD326+ intestinal epithelial cells (IECs) after allo-BMT resulted in decreased histone acetylation, which was restored after local administration of exogenous butyrate. Butyrate restoration improved IEC junctional integrity, decreased apoptosis and mitigated GVHD. Furthermore, alteration of the indigenous microbiota with 17 rationally selected strains of high butyrate–producing Clostridia also decreased GVHD. These data demonstrate a heretofore unrecognized role of microbial metabolites and suggest that local and specific alteration of microbial metabolites has direct salutary effects on GVHD target tissues and can mitigate disease severity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
26 August 2016
In the version of this article initially published, the middle initial (C) for the sixteenth author was incorrect. The correct author name should be Thomas M. Schmidt (and the corresponding initials in the Author Contributions section should be T.M.S.). The error has been corrected in the HTML and PDF versions of the article.
References
David, L.A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Turnbaugh, P.J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Mathewson, N. & Reddy, P. The microbiome and graft versus host disease. Current Stem Cell Reports 1, 39–47 (2015).
Jenq, R.R. et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 209, 903–911 (2012).
Eriguchi, Y. et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood 120, 223–231 (2012).
Jenq, R.R. & van den Brink, M.R.M. Allogeneic haematopoietic stem cell transplantation: individualized stem cell and immune therapy of cancer. Nat. Rev. Cancer 10, 213–221 (2010).
Choi, S. & Reddy, P. Graft-versus-host disease. Panminerva Med. 52, 111–124 (2010).
Ferrara, J.L.M., Levine, J.E., Reddy, P. & Holler, E. Graft-versus-host disease. Lancet 373, 1550–1561 (2009).
Taur, Y. et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 124, 1174–1182 (2014).
Ganapathy, V., Thangaraju, M., Prasad, P.D., Martin, P.M. & Singh, N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr. Opin. Pharmacol. 13, 869–874 (2013).
Fleming, L.L. & Floch, M.H. Digestion and absorption of fiber carbohydrate in the colon. Am. J. Gastroenterol. 81, 507–511 (1986).
Sealy, L. & Chalkley, R. The effect of sodium butyrate on histone modification. Cell 14, 115–121 (1978).
Cook, S.I. & Sellin, J.H. Review article: short chain fatty acids in health and disease. Aliment. Pharmacol. Ther. 12, 499–507 (1998).
Reddy, P. et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc. Natl. Acad. Sci. USA 101, 3921–3926 (2004).
Reddy, P. et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J. Clin. Invest. 118, 2562–2573 (2008).
Choi, S.W. et al. Vorinostat plus tacrolimus and mycophenolate to prevent graft-versus-host disease after related-donor reduced-intensity conditioning allogeneic haemopoietic stem-cell transplantation: a phase 1/2 trial. Lancet Oncol. 15, 87–95 (2014).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Hill, G.R. & Ferrara, J.L. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood 95, 2754–2759 (2000).
Gao, S.-M. et al. Histone deacetylases inhibitor sodium butyrate inhibits JAK2/STAT signaling through upregulation of SOCS1 and SOCS3 mediated by HDAC8 inhibition in myeloproliferative neoplasms. Exp. Hematol. 41, 261–270 (2013).
Ell, B. & Kang, Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 23, 603–611 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Charney, A.N., Micic, L. & Egnor, R.W. Nonionic diffusion of short-chain fatty acids across rat colon. Am. J. Physiol. 274, G518–G524 (1998).
Sun, Y. et al. Cutting edge: negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J. Immunol. 182, 5899–5903 (2009).
Noth, R. et al. Increased intestinal permeability and tight junction disruption by altered expression and localization of occludin in a murine graft versus host disease model. BMC Gastroenterol. 11, 109 (2011).
Soler, A.P. et al. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis 20, 1425–1431 (1999).
Suzuki, T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 70, 631–659 (2013).
Hanash, A.M. et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity 37, 339–350 (2012).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Chang, P.V., Hao, L., Offermanns, S. & Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 111, 2247–2252 (2014).
Lahl, K. et al. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 204, 57–63 (2007).
Lahl, K. & Sparwasser, T. In vivo depletion of FoxP3+ Tregs using the DEREG mouse model. Methods Mol. Biol. 707, 157–172 (2011).
Topham, C.H. & Taylor, S.S. Mitosis and apoptosis: how is the balance set? Curr. Opin. Cell Biol. 25, 780–785 (2013).
Wong, J.M.W., de Souza, R., Kendall, C.W.C., Emam, A. & Jenkins, D.J.A. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243 (2006).
Narushima, S. et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 5, 333–339 (2014).
Schloss, P.D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Schloss, P.D., Gevers, D. & Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6, e27310 (2011).
DeSantis, T.Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Cresci, G.A., Thangaraju, M., Mellinger, J.D., Liu, K. & Ganapathy, V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J. Gastrointest. Surg. 14, 449–461 (2010).
Takashima, S. et al. The Wnt agonist R-spondin1 regulates systemic graft-versus-host disease by protecting intestinal stem cells. J. Exp. Med. 208, 285–294 (2011).
Iyengar, S., Zhan, C., Lu, J., Korngold, R. & Schwartz, D.H. Treatment with a rho kinase inhibitor improves survival from graft-versus-host disease in mice after MHC-haploidentical hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 20, 1104–1111 (2014).
Lefrançois, L. & Lycke, N. Isolation of Mouse Small Intestinal Intraepithelial Lymphocytes, Peyer's Patch, and Lamina Propria Cells (John Wiley & Sons, 2001).
Reddy, P. et al. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat. Med. 11, 1244–1249 (2005).
Mathew, A.V., Seymour, E.M., Byun, J., Pennathur, S. & Hummel, S.L. Altered metabolic profile with sodium-restricted dietary approaches to stop hypertension diet in hypertensive heart failure with preserved ejection fraction. J. Card. Fail. 21, 963–967 (2015).
Mell, B. et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genomics 47, 187–197 (2015).
Koenigsknecht, M.J. et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect. Immun. 83, 934–941 (2015).
Cole, J.R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 (2009).
Wang, Q., Garrity, G.M., Tiedje, J.M. & Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Acknowledgements
This work was supported by the US National Institutes of Health (NIH) (National Heart, Lung and Blood Institute, grants HL-090775 and HL-128046 (to P.R.); National Cancer Institute, grant CA-173878 (to P.R.)). Metabolomic studies were performed through the Molecular Phenotyping Core, Michigan Nutrition and Obesity Center (grant DK089503 (to S.P.)) and Michigan Regional Metabolomics Resource Core (grant DK097153 (to S.P.)). We acknowledge use of the Microscopy & Image-analysis Laboratory (MIL) of the University of Michigan's Biomedical Research Core Facilities for preparation of samples and images. Support for the MIL core is provided by the University of Michigan Cancer Center (NIH grant CA46592) and the University of Michigan Gut Peptide Research Center (NIH grant DK34933).
Author information
Authors and Affiliations
Contributions
N.D.M. designed experiments, performed experiments, analyzed data and wrote the paper. A.V.M. and M.K. performed experiments and analyzed data. R.J. and A.H. designed and performed experiments. T.T., K.O.-W., S.-R.W., Y.S., C.R., H.F., J.B., Y.S., C.L. and M.C. performed experiments. T.M.S., V.B.Y. and M.v.d.B. wrote the paper. K.H. provided reagents and wrote the paper. S.P. analyzed data and wrote the paper. P.R. designed experiments, analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
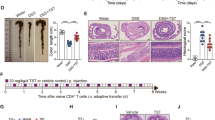
Supplementary Figure 1 Allogeneic BMT reduces amounts of intracellular butyrate in IECs.
(a) Schematic of fatty acid analysis. Animal cages of naive animals and those receiving syngeneic or allogeneic BMT were exchanged on day 3 relative to transplant. All groups were killed on day 7 and day 21 (Fig. S2). Homogenized samples were normalized and analyzed via gas chromatography mass spectrometry. Heatmap of fatty acid profiles (short and long chain) of all animals combined, that were killed 7 days following syngeneic (BALB/c → BALB/c) or allogeneic (C57BL/6J → BALB/c) transplant in the (b) intestinal luminal contents (stool) and (d) in the intestinal tissue. n = 9 – 10 animals per group. (c) Butyrate concentration was not different in the serum, spleen, and liver of naive animals or recipients of syngeneic or allogeneic BMT.
Supplementary Figure 2 Reduced amounts of intracellular butyrate in IECs after BMT.
(a) Fatty acid levels (short and long chain) 21 days following transplant in the (left) intestinal luminal contents (stool) and in the (right) intestinal tissue. Quantifications of the short chain fatty acids acetate, propionate, and butyrate are shown below representative heatmaps; n = 5 animals per group.
Supplementary Figure 3 Allo-BMT reduces histone acetylation and receptor expression in IECs.
(a) Protein expression of acetyl-histone H4 and densitometric analysis (below blots) normalized to presence of ɑ-tubulin in CD326+ purified intestinal epithelial cells (IECs) 7 days following syngeneic (BALB/c → BALB/c) or allogeneic (C57BL/6J → BALB/c) BMT. (b) Gene expression and (c) protein level of GPR43 in IECs (CD326+) of syngeneic or allogeneic BMT recipients 21 days following transplant. (d) Gene expression of Slc5a8 (left) and Gpr43 (right) in IECs (CD326+) 7 days following syngeneic or allogeneic BMT. Representative immunoblots; densitometric analysis of three similar experiments combined, are shown below blots. (e) Primary IECs (CD326+) cultured in the presence or absence of inflammatory cytokines IFN-γ (40ng/ml) or TNF (500ng/ml) for 6 hours. (f) Schema of butyrate entry and metabolism in to the intestinal epithelial cell. Butyrate enters the epithelium from the lumen and subsequently undergoes mitochondrial β-oxidation. The product acetyl-CoA enters the tricarboxylic cycle. The butyrate, citrate, and malate pools were sampled for this study to assess butyrate metabolic flux. (g)-(h) MFA showing 13C label incorporation in tricarboxylic acid metabolite pools of syngeneic and allogeneic mice fed a bolus (2 g/kg) of 13C2-Butyrate or non-labeled 12C-Butyrate, n=6 mice per group. (g) Percent incorporation of 13C in succinate of the large intestine tissue. (h) Left panel represents succinate and right panel represents malate 13C-label incorporation in small and large intestine tissues combined. ND = not detected.
Supplementary Figure 4 Intragastric butyrate increases survival of allo-BMT recipients.
(a) Improved survival was observed using a second model of syngeneic (C57BL/6J → C57BL/6J) and allogeneic (C3H.sw → C57BL/6J) BMT indicating strain independent effects. Each recipient received 2 x 106 T cells and 5 x 106 bone marrow cells following lethal irradiation (1000 cGy) on day -1; syngeneic n=6, n=12 per allogeneic group. (b) Liver and intestinal histopathology was decreased 21 days post-BMT in recipients of allogeneic BMT treated with intragastric butyrate in minor antigen mismatch model. (c) Significantly decreased weight loss (d) and significantly improved survival was observed in allogeneic recipients of parent into F1 BMT model (C57BL/6J → B6D2F1) that received butyrate treatment. Each recipient received 50 x 106 whole splenocytes and 5 x 106 bone marrow cells; syngeneic n=5, n=10 per allogeneic group. (e) Significantly improved survival was still observed in recipients of high donor allo-T cell dose (2.5 x 106) that were treated with intragastric butyrate (10 mg/kg). Each recipient received either syngeneic (C57BL/6J → C57BL/6J) or allogeneic (C57BL/6J → Balb/c) donor T cells and 5 x 106 bone marrow; syngeneic n=5, n=10 per allogeneic group. (f) Transmission electron microscopy (TEM) of intestines, isolated from recipients of syngeneic (BALB/c → BALB/c) or allogeneic (C57BL/6J → BALB/c) transplant with or without daily intragastric gavage of butyrate for duration of experiment; all samples were isolated 21 days following BMT and stained with ruthenium red (0.1%). Increased ruthenium red leakage observed in recipients of allogeneic transplant treated with vehicle compared to butyrate treatment or syngeneic BMT; arrows indicate cell-cell interface. (g) Total CD45+ intestinal cell numbers harvested were not different between recipients of allogeneic (C57BL/6J → BALB/c) BMT treated with butyrate or vehicle. (h) Treg cells were depleted by i.p. injection of diphtheria toxin (10 µg/kg) on day -2 and day -1 of donors prior to transplant. (i) Decreased weight loss in BMT recipients of Treg depleted (B6 DT.Treg) donor T cells (0.5 x 106) treated with butyrate. (j) HAT activity was not altered in T cells of the spleen 21 days following syngeneic or allogeneic BMT. All BMTs had n=10 per group.
Supplementary Figure 5 The IEC barrier is enhanced by butyrate treatment.
(a) Butyrate treatment (1mM) resulted in increased expression of junctional claudins (1, 5, 6, 10, 11, 13, 14, 17, and 18) in cultured primary intestinal organoids, compared to control. (b) Decreased gene expression of the pro-apoptotic proteins Bak1 (left) and Bax (right) and (c) increased expression of the junctional protein Occludin (Ocln) in IECs (CD326+) treated with vehicle or butyrate 1mM.
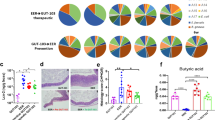
Supplementary Figure 6 Rationally altering the commensal microbiota.
(a) Detailed presence of bacterial strains found in animals colonized with 17-strain cocktail. 16S rRNA-encoding gene sequencing of stool for percent of 17 Clostridal strains in total GI community on day -1 and day +35, relative to allogeneic (C57BL/6J → BALB/c) BMT on day 0. Vehicle or 17-strain cocktail were administered to recipients every other day via intragastric gavage beginning day -14 and continued through day +21, relative to BMT. ND = not detected. (b) Antibiotic mixture (ampicillin 5 mg, metronidazole 4 mg, clindamycin 5 mg, and vancomycin 5 mg) was administered by intragastric gavage for 6 days to target obligate anaerobes of C57BL/6J (H-2b) mice. Untreated mice or mice that received antibiotic mixture, were used as recipients of allogeneic BMT (B10.BR → C57BL/6J).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 2 (PDF 2464 kb)
Supplementary Table 1: qPCR primers
Primers used with gene names indicated. (XLSX 30 kb)
Rights and permissions
About this article
Cite this article
Mathewson, N., Jenq, R., Mathew, A. et al. Gut microbiome–derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 17, 505–513 (2016). https://doi.org/10.1038/ni.3400
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3400
This article is cited by
-
Association of gut microbiota with the pathogenesis of SARS-CoV-2 Infection in people living with HIV
BMC Microbiology (2024)
-
Bacteria and bacteriophage consortia are associated with protective intestinal metabolites in patients receiving stem cell transplantation
Nature Cancer (2024)
-
Role of the microbiota in response to and recovery from cancer therapy
Nature Reviews Immunology (2023)
-
Feasibility of a dietary intervention to modify gut microbial metabolism in patients with hematopoietic stem cell transplantation
Nature Medicine (2023)
-
Gut OncoMicrobiome Signatures (GOMS) as next-generation biomarkers for cancer immunotherapy
Nature Reviews Clinical Oncology (2023)