Abstract
Endometrial stromal sarcomas represent the second most common mesenchymal uterine tumor. The 2003 WHO classification distinguishes low-grade and undifferentiated endometrial stromal sarcomas with different prognoses. Endometrial stromal sarcomas are a genetically heterogeneous group of sarcomas harboring different cytogenetic anomalies. Recently, a fusion between the YWHAE and FAM22A/B genes subsequent to a t(10;17) (q22;p13) has been described in endometrial sarcomas with high-grade histology. We examined YWHAE rearrangements by FISH break-apart and RT-PCR in a series of 27 undifferentiated uterine stromal sarcoma without JAZF1 rearrangements. Immunohistochemistry (IHC) was carried out with a panel of antibodies (estrogen (ER) and progesterone (PR) receptors, CD10, Cyclin D1, β-catenin, p53, and Ki-67). We identified a subgroup of endometrial sarcomas with high-grade histology and uniform morphology harboring YWHAE rearrangements. FISH break-apart was interpretable in 20 cases (74%). Twelve cases (60%) showed <10% of tumor cells with a YWHAE rearrangement, 4 cases (20%) showed between 10 and ≤20%, and 4 (20%) >20%. RT-PCR was tested on 24/27 cases (88%) and 19 cases were interpretable (79%). Five cases (26%) showed a specific fusion transcript YWHAE–FAM22A/B sequence. The best concordance rate between FISH and RT-PCR (94%) was obtained with the threshold of 20% of cells with a YWHAE rearrangement. The YWHAE-rearranged cases showed high-grade morphology with uniform appearance, spindle or round epithelioid cells, low ER and PR, CD10 expression, and a high and diffuse positivity for Cyclin D1, p53, and nuclear β-catenin negativity. Cyclin D1 was the most sensitive marker for high-grade endometrial sarcomas with YWHAE rearrangement. All undifferentiated uterine sarcomas with pleomorphic appearances did not harbor any YWHAE rearrangements, except for one case. Overall, for endometrial sarcoma cases with high-grade morphology we recommend to test for YWHAE rearrangements by FISH break-apart, a cost- and time-efficient method, and to complete the investigation by RT-PCR in borderline cases.
Similar content being viewed by others
Main
Endometrial stromal sarcomas are malignant uterine mesenchymal tumors that represent <10% of uterine mesenchymal neoplasms and 1% of all uterine cancers.1, 2 They are the second most common mesenchymal tumor of the uterus, developing especially in perimenopausal women.1
According to the 2003 WHO classification, endometrial stromal sarcomas are divided into low-grade and undifferentiated, depending on tumor morphology and irrespective of tumor mitotic count. Indeed, low-grade endometrial stromal sarcomas are characterized by small oval or fusiform (blue) cells appearing similar to proliferative endometrial stroma with (often extensive) myometrial permeation, frequent lymphatic invasion, and indolent clinical behavior.3 Undifferentiated endometrial stromal sarcomas, previously described as high-grade endometrial stromal sarcomas are a group of undifferentiated tumors bearing little or no resemblance to low-grade with frequent necrosis, destructive myometrial invasion, pleomorphic nuclei, and brisk mitotic activity.4 Distinction between these two categories of tumors has a crucial clinical impact because their prognosis is very different.5, 6
Endometrial stromal sarcomas represent a genetically heterogeneous group of uterine sarcomas. Almost 60% of low-grade show t(7;17)(p15;q12) rearrangements involving JAZF1-SUZ12, 1% JAZF1-PHF1 t(6;7)(p21;p15), and about <1% EPC1-PHF1 t(6;10;10)(p21;q22;p11).7 These genes belong to a family of genes involved in chromatin binding. Recently, Lee et al.8 identified the genes rearranged in the t(10;17)(q22;p13) translocation previously described in endometrial stromal sarcoma9, 10, 11 and also in pediatric clear cell sarcoma of the kidney,12, 13 corresponding to an in-frame YWHAE-FAM22 fusion of YWHAE exons 5–2 of FAM22A or FAM22B.8
The goal of the present study was to analyze the rearrangement of the YWHAE gene by FISH break-apart and RT-PCR techniques, and to identify correlations between YWHAE gene status and morphology, immunohistochemistry (IHC), and clinical behavior in a series of high-grade-morphology endometrial stromal sarcomas with no JAZF1 rearrangements. Given the lack of published data about YWHAE FISH break-apart,8 we also aimed to identify the optimal threshold for positivity in RT-PCR for cell rearrangements.
Materials and methods
Tissue Samples
A total of 27 cases of high-grade histology endometrial stromal sarcomas with no JAZF1 rearrangements diagnosed between 1995 and 2011, as undifferentiated endometrial stromal sarcomas were retrospectively identified from the prospectively maintained Department of Pathology records at the Institut Bergonié of Bordeaux. Tumoral blocks for 24 cases with a total of 35 blocks were available. For the remaining three cases, only the slides were available. Of the 27 cases, 23 were fixed in neutral buffered formalin and 4 in Bouin. Hematoxylin and eosin-stained slides were reviewed by two pathologists (SC and GMG) and cases were diagnosed according to the 2003 WHO classification and, in line with Kurihara et al’s14 paper, separated into uniform and pleomorphic types. Eight of the 27 cases were pleomorphic types (or true undifferentiated according to Lee et al.15) and 19 were uniform types, showing monomorphic spindle, fibromyxoid, or epithelioid round cells. Clinical data (age, stage, and outcome) were available for 24/27 patients. The series included 25 primary uterine endometrial stromal sarcomas and two para-uterine tumors.
The samples used in this study as part of the Biological Resources Center of the Institut Bergonié, Comprehensive Cancer Centre. In accordance with the French Public Health Code (articles L. 1243-4 and R. 1243-61), the Biological Resources Center of Institut Bergonié has received agreement from the French authorities to deliver samples for scientific research (number AC-2008-812), approved by the Committee of Protection of Individuals.
FISH Probe Design and Development
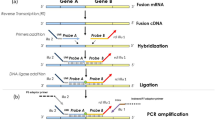
In-house FISH probes directed to centromeric and telomeric ends of YWHAE gene according to a UCSC genome browser were cloned using a bacterial artificial chromosome (BAC). The following BAC clones were employed after verification on chromosomal spread: telomeric CTD-2507J6 (190 kb), RP11-356I18 (172 kb), and RP11-1061C17 (216 kb) labeled with SpectrumOrange-dUTP (Abbott) and centromeric RP11-100F18 (19 918 kb) and CTD-3249J3 (180 kb) labeled with 5(6)-SpectrumGreen-dUTP (Abbott) (Figure 1a).
Sample Preparation, Hybridization, and Detection
FISH assay was performed using the Histology FISH accessory kit (Dakopatts, Denmark). Briefly, sections from representative blocks were cut onto positively charged slides at 5 μm thickness. Deparaffinization of the sections was carried out with three 10-min immersions in xylene, followed by two 3-min immersions in wash buffer. DNA was retrieved by immersing the slides in MES (2-[N-morpholino]ethanesulphonic acid) buffer for 10 min at 97 °C, then by slowly cooling to room temperature. Slides were then subjected to pepsin digestion for 7 min at 37 °C, dehydrated, and air-dried. Codenaturation (82 °C) and hybridization (37 °C) were achieved by placing the slides into the Hybridizer (Dakopatts). The next day, the slides were washed in stringent buffer for 10 min at 65 °C and nuclei were counterstained by using DAPI/antifade solution.
FISH Analysis and Scoring
Green and orange fluorescent signals were counted in 100 tumor nuclei under a Nikon Eclipse 80i fluorescent microscope with appropriate filters, and pictures were captured using a Hamamatsu C4742-95 CCD camera and analyzed with the Genikon software (Alphelys, France). Paired signals were defined as a single yellow (overlapping) signal or a red and green signal <2 signal diameters apart, whereas unpaired signals were those separated by >2 signal diameters from an oppositely colored signal. For each case, 100 nuclei were counted and a rearrangement was defined when the cell displayed one fusion, one green, and one orange signal pattern.
To identify the best threshold of the cells with rearrangement by comparison with the RT-PCR results, we first set the percentage of cells with rearrangement and then tested the positivity threshold at 10, 20, and 30% of cells with rearrangement.
Real-Time Reverse Transcription-Polymerase Chain Reaction Analysis for YWHAE–FAM22A/B Fusion Gene
RNA extraction
We selected the tumor areas for core sampling on the same fields analyzed by FISH.
After being deparaffinized, sections were digested for 3 days at 55 °C with proteinase K (Qiagen, Courtaboeuf, France) (final concentration was 4 mg/ml). RNA were extracted with Trizol-LS reagent (Invitrogen, Cergy Pontoise, France) according to the manufacturer’s instructions. RNA pellets resuspended in 10 μl RNase-free water were incubated with Mix 1X (Promega, Madison WI, USA), dithiothreitol 1 mM (Roche), Rnasin 40U (Promega), and DNase 2U (Promega) for 1 h at 42 °C. Then, RNA were repurified using a Trizol-LS reagent.
Real-time RT-PCR and sequencing
Reverse transcription of 5 μg RNA was performed in a total volume of 20 μl with 50 mM Tris-HCl, pH 8.3, 40 mM KCl, 5 mM MgCl2, 0.5% Tween, 0.5 mM dNTP mix, 10 mM dithiothreitol, specific reverse primer (FAM22 or reverse primer β2-microglobulin), 12 U RNAse inhibitor (Promega), and 10U Expand Reverse Transcriptase (Roche Diagnostics, Meylan, France). Samples were incubated at 42 °C for 1 h, then at 95 °C for 5 min.
PCR amplification was performed in duplicate using a 96-well plate (Applied Biosystems, Foster City, CA, USA) with a 50-μl final reaction mixture containing: 300 nM of each primer, 200, and 50 nM, respectively, of probe FAM22 and probe β2-microglobulin, and 0.25 U of Amperase UNG in a 2 × qPCR Mastermix plus-low Rox (Eurogentec, Herstal, Belgium). Thermal cycling conditions were 2 min at 50 °C for Amperase activation, 10 min at 95 °C for Taq polymerase activation, then 50 cycles of two PCR steps consisting of 30 s at 95 °C, and 1 min at 60 °C. All reactions were performed in the ABI Prism 7500 Sequence Detection System (Applied Biosystems).
Primers (YWHAE+5′ CACTTATCATGCAGTTGTTACGTGAT-3′; FAM22-5′-GGGCAGAGCCGTGAACAC-3′; Beta2m+5′-TGACTTTGTCACAGCCCAAGATA-3′; Beta2m- 5′ AATCCAAATGCGGCATCTTC-3′); and probe sequences (FAM22 5′-FAM-ACCGGGCGTGACCGCGA-TAMRA-3′and Beta2m 5′-FAM-TGATGCTGCTTACATGTCTCGATCCCA-TAMRA-3′) were chosen with the software Primer Express (Applied Biosystems) (Figure 1b). Primers were purchased from Eurofins MWG Operon (Ebersberg, Germany) and probes, dNTP, and Amperase UNG were purchased from Eurogentec.
For sequencing, purification of the PCR product was performed using the GFX PCR DNA and gel band purification kit (GE Healthcare Lifesciences, Buckinghamshire, UK). Automated cycle sequencing for both strands (with the YWHAE+ or FAM22− primers) was performed using the Bid-Dye DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). Sequencing reactions were carried out on the ABI Prism 310 Genetic Analyzer (Applied Biosystems). Sequences were compared to the normal sequence with the SeqScape v2.5 software (Applied Biosystems).
Immunohistochemistry
Representative paraffin-embedded sections were available for the IHC study for 25/27 cases. We performed a panel of antibodies: estrogen receptor (ER), progesterone receptor (PR), desmin, H-caldesmon, CD10, Cyclin D1, β-catenin, p53 and Ki-67. Difficult cases were investigated with additional antibodies (AE1–AE3 Cytokeratin, PS100, MDM2, calretinin, HMB-45, and Melan A) at diagnosis to exclude other diagnostic hypotheses. The heat-induced antigen retrieval was performed using the ULTRA Cell Ventana conditioning buffer (CC1), pH 8 (EDTA) at 95 °C. UltraView Ventana was the DAB polymer detection complex used. All steps were performed in the Roche/Ventana Benchmark Ultra automat (For primary antibody specifications and staining conditions, see Table 1).
For Ki-67, p53, ER, PR, β-catenin, and Cyclin D1, only the nuclear staining was considered positive. For p53 staining, we scored cases: 0 (0% of nuclei stained), 1 (<5% of nuclei stained), 2 (5–25% of nuclei stained), 3 (26–50% of nuclei stained), 4 (51–75% of nuclei stained), 5 (76–95% of nuclei stained), or 6 (>95% of nuclei stained).16, 17 According to current knowledge about p53 staining in many organs (for example, ovary, esophagus, or endometrium) as a phenomenon of all or nothing,18 we considered as positive (p53-positive) the p53 scores from 4 (51–75% of positive nuclei) to 6 (>95% of positive nuclei). Cases with scores from 1 to 3 were considered as negative (p53-wild type). Cases with no staining scores of 0 (p53-absent), so-called null patterns, were considered potentially related to P53 mutation (nonsense, frameshift, or splice site).18 For Cyclin D1, we chose a cutof ≥70% of cells with strong nuclear staining.16, 18
Results
FISH and RT-PCR Results, and Correlations
(Table 2) Seven cases (26%) were not interpretable because of fixation or extensive tumoral necrosis. Twelve cases (60%) showed <10% of tumor cells with YWHAE rearrangement; four (20%) showed ≥10% but ≤20% of cells with rearrangement, and four (20%) showed >20% of cells with rearrangement (Table 2).
RT-PCR was tested on 24/27 available blocks. The mRNA quality was insufficient for analyses in 3/24 cases (12%) due to significant degradation of mRNA in paraffin-embedded tumors. Among 19 interpretable cases, 5 (26%) showed a specific fusion transcript that was sequenced; 14/19 (74%) were negative. Among the 12 FISH-negative cases, one (8%) showed a specific fusion transcript by RT-PCR. Of the four FISH-positive cases (>20% of cells with rearrangement), one case was not interpretable and three (75%) were positive by RT-PCR. Of the four cases with between 10 and ≤20% of cells with rearrangement (uncertain cases), three cases were RT-PCR-negative, and one block was not available (Table 2).
With a threshold of 10% tumor cells with YWHAE rearrangements, 8/20(40%) cases in the series were considered positive. Of these eight cases, three YWHAE-positive cases (40%) showed specific transcript fusion at RT-PCR (concordant results), three (40%) did not show the specific transcript fusion (discordant), in one case RT-PCR was not interpretable, and for another case the block was not available. However, in our series, the best concordance rate (94%) between FISH and RT-PCR was achieved with the FISH threshold of 20%. With this cutoff, only four cases resulted FISH-positive (20%), with specific transcript fusion revealed in three cases (in one case RT-PCR was not interpretable). There was no significant difference with a threshold of 30%, therefore, we decided to adopt a ‘cautious’ threshold of 30%.
Histopathological and Immunophenotypical Features of ESS
All but one YWHAE-rearranged case (established at the 30% threshold) showed common morphologic features: a uniform spindle, fibromyxoid, or round-cell epithelioid appearance often arranged in nests with an intermediate grade of atypia between the low-grade endometrial stromal sarcomas and the pleomorphic types (Figure 2). One YWHAE-rearranged case showed purely spindle cell appearances (cases 27) and one case a pure fibromyxoid feature (case 9). Among the other YWHAE-rearranged cases, two were mixed spindle and epithelioid round cells (cases 26 and 14), one case was pure epithelioid round cells (case 11), and one case (case 2) was of pleomorphic type (Table 2; Figure 3). Only one case (case 2), a discordant case that was FISH-negative, but RT-PCR-positive, was morphologically different from the other YWHAE-rearranged endometrial stromal sarcomas and showed pleomorphic morphology with high-grade nuclear atypia, prominent nucleoli, and no resemblance to endometrial stroma (Figure 4). Among the groups of YWHAE-rearranged cases, 4/6 showed a ‘tongue-like’ growth pattern, as seen in low-grade endometrial stromal sarcomas (Table 2).
Morphological spectrum of undifferentiated endometrial stromal sarcoma uniform type (high-grade endometrial stromal sarcoma). A case with spindle cell features arranged in uniform sheets (H&E × 100 PF) (a). High mitotic rate is frequent in high-grade tumors (H&E × 400 HP) (b). Fibromyxoid appearance was the dominant component in two cases. A myxoid background crossed by thin vessels (H&E × 100 HP) (c) surrounds spindle, and round cells with vacuolated nuclei (H&E × 400 HP) (d).
Morphological, immunophenotypic, and genotypic features of a YWHAE-rearranged endometrial stromal sarcoma (case 26). Undifferentiated endometrial stromal sarcoma uniform type (high grade) with diffuse myometrial permeative infiltration and lymphovascular involvement (H&E × 20 PF) (a). The tumor is composed of uniform spindle cells with elongated nuclei showing angulated contours (H&E × 200 PF) (b), with strong and diffuse Cyclin D1 immunoreactivity (c), and YWHAE FISH break-apart rearrangement (d). Detection by RT-PCR of the fusion transcript sequenced (e).
Morphological, immunophenotypic, and genotypic features of a undifferentiated endometrial stromal sarcoma pleomorphic type, a poorly differentiated sarcoma (case 6). The tumor shows a diffuse architecture with destructive infiltration of surrounding tissue (a). The cells are not similar in appearance to proliferative-phase endometrial stroma but show high-grade nuclear atypia, often with giant cell pleomorphic appearance (b). p53 is highly expressed (c), and Cyclin D1 is negative (d). CD10 is variably and weakly expressed (e). No YWHAE rearrangement is detected by FISH break-apart (f).
At the immunohistochemical level, all YWHAE-rearranged cases, with the exception of the pleomorphic one (case 2), had the same pattern: Cyclin D1 strongly positively expressed by 100% of the YWHAE-rearranged cases (FISH+), with p53 and β-catenin negativity. Taking the RT-PCR or FISH-positive cases, Cyclin D1 was positive in 4/6 cases. The two negatives cases were (case 2) a pleomorphic type p53-positive and a RT-PCR-positive case, not interpretable by FISH (case 11). On the other hand, among the Cyclin D1-positive cases, all (7/7) were uniform types. From all of the Cyclin D1-positive cases, 4/7 were YWHAE-rearranged, two were not interpretable via FISH analysis, and one was YWHAE-FISH-negative (Table 2). No nuclear staining for β-catenin was detected except in one case (case 16, a uniform ESS with non-rearranged YWHAE spindle cells). CD10 was negative in 3/4 cases, ER was negative in all cases (4/4), and PR was positive in one case (1/4) (Table 2). The YWHAE-FISH-negative but RT-PCR-positive case (case 2) showed strong expression of p53 and CD10, and no immunostaining for Cyclin D1, ER, and PR.
p53 immunostaining in pleomorphic type sarcomas showed both a pattern of protein accumulation (p53-positive) and a pattern of complete absence of staining (p53-absent) (Table 2).
Clinicopathological Correlations and YWHAE Status
The 27 patients ranged in age from 31 to 88 years, with a median age of 59 years. Stage (FIGO 2009) was available for 22: 14 (64%) patients were stage IB, 1 (4%) stage IIA, 1 (4%) stage IIB, 1 (4%) stage IIIB, 2 (10%) stage IIIC, and 3 (14%) stage IVB. Clinical follow-up was available for 25/27, and for the patients who were alive during follow-up it varied from 13 months to 10 years after the initial diagnosis. Of these 25 patients with clinical follow-up, 13 (52%) died of the disease, 3 (12%)died of unknown causes, 4 (16%) were alive without evidence of disease at the last follow-up, 4 (16%) were alive with disease, and detailed follow-up information was not available for the remaining patient (4%); we only know that she was alive (Table 2). Among the YWHAE-positive cases (FISH or RT-PCR), one patient had died of disease, one was alive without evidence of disease, two alive with disease, one alive but disease status was unknown, and one patient was lost to follow-up. Among the YWHAE-negative cases, nine patients had died of disease, one deceased of unknown causes, two alive without evidence of disease, and two alive with disease. The small number of events in each group does not allow us to conclude a significant survival difference between the two tumor groups.
Eight cases overall (30%) showed pleomorphic morphology with high-grade nuclear atypia, prominent nucleoli, and no resemblance to endometrial stroma. One case showed multinucleated giant osteoclastic-like cells. The myometrial invasion pattern was of the destructive and not-infiltrative type (as in low-grade endometrial stromal sarcoma or uniform-type undifferentiated endometrial stromal sarcoma). The median mitotic count for pleomorphic-type endometrial stromal sarcoma was 28 mitosis/10 HPF (14.8/mm2). Among the eight pleomorphic tumors, eight had died of disease.
Nineteen cases showed uniform appearance (70%) and among them, eight contained a composite pattern with a mixture of spindle cell areas and fibromyxoid or spindle, and epithelioid round cells, often arranged in nests with an intermediate atypia grade between the low-grade endometrial stromal sarcoma and the pleomorphic-type cells. The median mitotic count for uniform-type sarcoma was 18 mitosis/10 HPF (9.2/mm2). Clinical follow-up information was available for 17/19, as shown by the morphological and clinical features in Table 2. Endometrial stromal sarcoma is an aggressive disease with unfavorable behavior. Although the follow-up results are not statistically significant because of small number of events, the YWHAE-rearranged endometrial stromal sarcomas show better prognosis (Table 2) compared with tumors without YWHAE rearrangements.
It should be noted that, among the groups of patients who died of disease, the FIGO stages were advanced (stage III–IV) in six patients (40%) and were low (stage I–II) in eight patients (60%). It was not possible to correlate these data with pleomorphic or uniform morphology, nor with YWHAE status, as the study group was too small.
Discussion
Endometrial stromal sarcomas are the second most common category of malignant mesenchymal tumors of the uterus.3 The 2003 WHO classification distinguishes low-grade endometrial stromal sarcomas composed of neoplastic cells that appear similar to the endometrial stromal cells of proliferative endometrium and undifferentiated ones that lack specific differentiation and do not bear histological resemblances to endometrial stroma.4 The distinction between these two categories is critical because of prognosis6 (85% 5-year survival for low grade compared with <50% for undifferentiated). Among these paradigmatic extremes, there are many variant morphologies that can complicate the diagnosis. In routine practice it is frequent to observe intermediate pattern/morphology between low grade and high grade, and it is not rare to observe areas of undifferentiated endometrial stromal sarcoma juxtaposed to low-grade endometrial stromal sarcoma.
Recently, Lee et al.8 have identified the genes rearranged in t(10;17)(q22;p13) resulting in an in-frame fusion between YWHAE (exon 1–5) and one of the two homologous genes FAM22A and FAM22B (exon 2–7). This rearrangement has been previously reported in some cases of undifferentiated endometrial stromal sarcomas of spindle and fibromyxoid morphology.9, 10, 11 YWHAE encodes for a protein of the 14-3-3 protein family that regulates signal transduction, cytoskeletal configuration, metabolism, differentiation, survival, and transcription in eukaryotic cells.19 The translocation-mediated YWHAE–FAM22A/B fusion is an oncogenic mechanism supporting/driving the undifferentiated endometrial stromal sarcoma cancerogenesis.8 Interestingly, YWHAE rearrangements have not been found in other gynecologic and non-gynecologic neoplasms (leiomyoma, leiomyosarcoma, adenosarcoma, and carcinosarcoma), except for clear cell sarcoma of the kidney.13
In this study, we analyzed the rearrangement of the YWHAE gene by FISH break-apart and RT-PCR in 27 cases of JAZF1 non-rearranged undifferentiated endometrial stromal sarcomas, using formalin- or Bouin-fixed and paraffin-embedded specimens. We demonstrate the feasibility of FISH break-apart detection in routine samples, as did Lee et al.15 with 13 high-grade endometrial stromal sarcoma, and we compare FISH and RT-PCR results. FISH break-apart shows a break in a target gene, highlighted by the unpaired green and red signals flanking the gene, irrespective of the partner (known or unknown). On the other hand, RT-PCR shows a specific fusion transcript produced by rearrangement of two (or more) genes. Although these methods use different approaches, we presupposed that the concordance of the RT-PCR and FISH break-apart results could help to determine the best threshold for proportion of rearranged cells, at which positivity should be defined.
For YWHAE FISH break-apart interpretation, the first issue was to determine the best threshold of cells with rearrangements. As no publications discussing YWHAE FISH were available when we started this study, we initially described the percentage of cells with rearrangements and we compared this with the RT-PCR results. We thus tested the cutoff of 10% for ‘positive’ FISH, as is used for other break-apart probes, and the thresholds of 20 and 30%. Recently, Lee et al.15 used 30% of cells with rearrangement as FISH threshold. In our series, no difference was seen between a cutoff of 20 or 30%.
The only discordant case when the threshold was 20% was a pleomorphic undifferentiated endometrial stromal sarcoma, with 0% of cells having rearrangement but a specific fusion transcript sequenced. Morphologically, this case was an epithelioid and pleomorphic undifferentiated endometrial stromal sarcoma with extensive necrosis in the advanced stage with bilateral pulmonary metastasis as diagnosis. The patient died of disease 6 months later. The tumor showed strong positivity for CD10, negativity for desmin, H-caldesmon and smooth muscle actin, ER, and PR. p53 staining showed positivity in >50% of the cells (score 4) and cyclin D1 was negative. A possible biological explanation of this discordance may be the physical mechanisms in the chromosomal rearrangement, with the insertion of a small portion of the YWHAE gene sufficient to produce a fusion transcript but too small to be detected by FISH (cryptic translocation). Another mechanism that could be evoked is the transplicing of the pre-mRNA for YWHAE–FAM22 genes. This mechanism was suggested by Li et al.20 to explain JAZF1-SUZ12 FISH-negative but RT-PCR-positive results with a transient and chimeric RNA in early proliferative or late secretory normal endometrial cells.
The question arises of whether the discordance between FISH break-apart and RT-PCR results (FISH+ and RT-PCR−) with the 10% threshold reflects a true biological reality or not. As earlier explained, more partners (some unknown) could be involved in the translocation. A dual fusion FISH could solve the problem. If the FISH dual fusion, targeting YWHAE as a partner and FAM22 as other partner, is negative, YWHAE is rearranged with another until now unknown partner. In this case, FISH break-apart would be more sensitive than FISH dual fusion or RT-PCR, but less specific as explained below.
Although the cautious threshold for FISH positivity of 30% of cells with rearrangements seems to be the reference cutoff, the biological significance of some cells (up to 20%) with YWHAE rearrangements is still unexplained. RT-PCR provided a result when FISH was not interpretable in three cases (in one case a specific sequenced fusion transcript was demonstrated, and two cases were negative). In two cases, neither FISH nor RT-PCR were interpretable, and for one case FISH was positive and RT-PCR not interpretable (Table 2).
In our opinion, these two techniques are complementary and it could be useful to search for YWHAE rearrangements by FISH break-apart that provides a result within a few days. If the FISH result is not interpretable or borderline (showing a percentage of cells with rearrangements between 10 and 30%), we then recommend the use of RT-PCR (with results within a week). Thus, YWHAE FISH break-apart represents a cost- and time-efficient method attainable on formalin-fixed and paraffin-embedded samples.
In this study, we identified a subgroup of endometrial stromal sarcomas with morphological, immunophenotypical, and genotypical characteristics (YWHAE gene rearrangements). At the morphological level, the YWHAE-rearranged endometrial stromal sarcomas of all of our series but one (case 2) showed uniform spindle, fibromyxoid, or epithelioid appearance in contrast to undifferentiated endometrial stromal sarcomas pleomorphic types. They showed permeative myometrial involvement with lymphovascular emboli, and no destructive involvement in contrast to pleomorphic undifferentiated endometrial stromal sarcomas. In 4/6 YWHAE-rearranged cases, the tumoral growth pattern was ‘tongue-like’, as described in low-grade endometrial stromal sarcomas, suggesting a possible transformation of a low-grade sarcoma into a high-grade more atypical and aggressive tumor. These tumors were distinguished from low-grade endometrial stromal sarcoma because of high-grade nuclear features (1–3 variations in nuclear size), high mitotic activity, ER and PR negativity, or weak and focal expression, and Cyclin D1 strong positivity as described by Lee et al.16 On the other hand, they were different from undifferentiated pleomorphic uterine sarcomas as they were less atypical, and had no massive and destructive growth patterns. At the immunohistochemical levels the pleomorphic undifferentiated sarcomas did not express Cyclin D1.
Immunophenotypically, the high-grade endometrial stromal sarcomas highly expressed Cyclin D1 (>70% of the nuclei) without p53 nuclear accumulation. They rarely expressed CD10 (a typical marker of stromal tumors), and ER and PR were negative or weakly expressed. On the other hand, the diagnosis of pleomorphic undifferentiated endometrial stromal sarcomas should be considered only after exclusion of poorly differentiated carcinomas, leiomyosarcomas, Mullerian mixed tumors, or a rare case of other undifferentiated sarcomas. Indeed Cyclin D1 is the more sensitive immunohistochemical marker for high-grade endometrial stromal sarcomas with YWHAE rearrangements. Even though our series does not enable us to carry out statistically valid between-group comparisons of prognosis, the YWHAE-rearranged sarcomas seem to have a better prognosis than the non-rearranged ones (Table 2).
In conclusion, we set up a tool that is feasible in routine practice to establish the diagnosis of undifferentiated uniform or high-grade endometrial stromal sarcoma types. When a uterine or gynecological tumor shows an intermediate morphological appearance between low-grade and high-grade without pleomorphic nuclei, and when endometrial stromal sarcoma has no JAZF1 rearrangements, a diagnosis of high-grade endometrial stromal sarcoma should be considered and YWHAE rearrangements should be tested by FISH or RT-PCR to support the diagnosis.
References
Koivisto-Korander R, Martinsen JI, Weiderpass E et al. Incidence of uterine leiomyosarcoma and endometrial stromal sarcoma in Nordic countries: results from NORDCAN and NOCCA databases. Maturitas 2012;72:56–60.
Norris HJ, Taylor HB . Mesenchymal tumors of the uterus. I. A clinical and pathological study of 53 endometrial stromal tumors. Cancer 1966;19:755–766.
Abeler VM, Royne O, Thoresen S et al. Uterine sarcomas in Norway. A histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology 2009;54:355–364.
Hendrickson MR, Tavassoli FA, McCluggage WG et al. Mesenchymal tumors and related lesions In: Tavassoli FA, Devilee P eds. Tumours of the Breast and Female Genital Organs. IARC Press: Lyon, France, 2003, pp 233–244.
D’Angelo E, Spagnoli LG, Prat J . Comparative clinicopathologic and immunohistochemical analysis of uterine sarcomas diagnosed using the World Health Organization classification system. Hum Pathol 2009;40:1571–1585.
Bartosch C, Exposito MI, Lopes JM . Low-grade endometrial stromal sarcoma and undifferentiated endometrial sarcoma: a comparative analysis emphasizing the importance of distinguishing between these two groups. Int J Surg Pathol 2010;18:286–291.
Chiang S, Ali R, Melnyk N et al. Frequency of known gene rearrangements in endometrial stromal tumors. Am J Surg Pathol 2011;35:1364–1372.
Lee CH, Ou WB, Marino-Enriquez A et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci USA 2012;109:929–934.
Amant F, Tousseyn T, Coenegrachts L et al. Case report of a poorly differentiated uterine tumour with t(10;17) translocation and neuroectodermal phenotype. Anticancer Res 2011;31:2367–2371.
Leunen K, Amant F, biec-Rychter M et al. Endometrial stromal sarcoma presenting as postpartum haemorrhage: report of a case with a sole t(10;17)(q22;p13) translocation. Gynecol Oncol 2003;91:265–271.
Regauer S, Emberger W, Reich O et al. Cytogenetic analyses of two new cases of endometrial stromal sarcoma-non-random reciprocal translocation t(10;17)(q22;p13) correlates with fibrous ESS. Histopathology 2008;52:780–783.
Punnett HH, Halligan GE, Zaeri N et al. Translocation 10;17 in clear cell sarcoma of the kidney. A first report. Cancer Genet Cytogenet 1989;41:123–128.
O'Meara E, Stack D, Lee CH et al. Characterization of the chromosomal translocation t(10;17)(q22;p13) in clear cell sarcoma of kidney. J Pathol 2012;227:72–80.
Kurihara S, Oda Y, Ohishi Y et al. Endometrial stromal sarcomas and related high-grade sarcomas: immunohistochemical and molecular genetic study of 31 cases. Am J Surg Pathol 2008;32:1228–1238.
Lee CH, Marino-Enriquez A, Ou W et al. The clinicopathologic features of YWHAE-FAM22 endometrial stromal sarcomas: a histologically high-grade and clinically aggressive tumor. Am J Surg Pathol 2012;36:641–653.
Lee CH, Ali RH, Rouzbahman M et al. Cyclin D1 as a diagnostic immunomarker for endometrial stromal sarcoma with YWHAE-FAM22 rearrangement. Am J Surg Pathol 2012;36:1562–1570.
McCluggage WG, Connolly LE, McGregor G et al. A Strategy for defining biologically relevant levels of p53 protein expression in clinical samples with reference to endometrial neoplasia. Int J Gynecol Pathol 2005;24:307–312.
McCluggage WG, Soslow RA, Gilks CB . Patterns of p53 immunoreactivity in endometrial carcinomas: 'all or nothing' staining is of importance. Histopathology 2011;59:786–803.
Mackintosh C . Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J 2004;381:329–342.
Li H, Wang J, Mor G et al. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 2008;321:1357–1361.
Acknowledgements
This work was supported by grants ARC and the Fondation de France. We acknowledge Pippa McKelvie-Sebileau and Dorothée Quincy of Institut Bergonié for manuscript correction.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Croce, S., Hostein, I., Ribeiro, A. et al. YWHAE rearrangement identified by FISH and RT-PCR in endometrial stromal sarcomas: genetic and pathological correlations. Mod Pathol 26, 1390–1400 (2013). https://doi.org/10.1038/modpathol.2013.69
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.69
Keywords
This article is cited by
-
Prognostic factors in undifferentiated uterine sarcoma: a subanalysis of the SARCUT study
Archives of Gynecology and Obstetrics (2023)
-
Endometriale und weitere seltene uterine Sarkome
Der Pathologe (2022)
-
Targeted RNA expression profiling identifies high-grade endometrial stromal sarcoma as a clinically relevant molecular subtype of uterine sarcoma
Modern Pathology (2021)
-
Recurrent uterine tumors resembling ovarian sex-cord tumors with the growth regulation by estrogen in breast cancer 1-nuclear receptor coactivator 2 fusion gene: a case report and literature review
Diagnostic Pathology (2020)
-
High-grade transformation of low-grade endometrial stromal sarcomas lacking YWHAE and BCOR genetic abnormalities
Modern Pathology (2020)