Abstract
High-grade endometrial stromal sarcoma (HGESS) may harbor YWHAE-NUTM2A/B fusion, ZC3H7B-BCOR fusion, and BCOR internal tandem duplication (ITD). NTRK3 upregulation and pan-Trk expression were reported in soft tissue lesions that share similar morphology and genetic abnormalities. To confirm these findings in HGESS, differential expression analysis was performed at gene level comparing 11 HGESS with 48 other uterine sarcomas, including 9 low-grade endometrial stromal sarcomas, 23 undifferentiated uterine sarcomas, and 16 leiomyosarcomas, using targeted RNA sequencing data. Pan-Trk immunohistochemistry was performed on 35 HGESS, including 10 tumors with RNA expression data, with genotypes previously confirmed by targeted RNA sequencing, fluorescence in situ hybridization, and/or genomic PCR. Unsupervised hierarchical clustering of the top 25% of differentially expressed probes identified three molecular groups: (1) high NTRK3, FGFR3, RET, BCOR, GLI1, and PTCH1 and low ESR1 expression; (2) low NTRK3, FGFR3, RET, BCOR, GLI1, and PTCH1 and high ESR1 expression; and (3) low NTRK3, FGFR3, RET, BCOR, GLI1, PTCH1, and ESR1 expression. Among HGESS, 64% of tumors clustered in group 1, while 27% clustered in group 2. Cytoplasmic and/or nuclear pan-Trk staining of variable extent and intensity was seen in 91% of HGESS regardless of cyclin D1 and/or BCOR positivity. ER and PR expression was seen in 44% of HGESS despite ESR1 downregulation. Two patients with ER and PR positive but ESR1 downregulated stage I HGESS were treated with endocrine therapy, and both recurred at 12 and 36 months after primary resection. By RNA expression, HGESS appear homogenous and distinct from other uterine sarcomas by activation of kinases, including NTRK3, and sonic hedgehog pathway genes along with downregulation of ESR1. Most HGESS demonstrate pan-Trk staining which may serve as a diagnostic biomarker. ESR1 downregulation is seen in some HGESS that express ER and PR which raises implications in the utility of endocrine therapy in these patients.
Similar content being viewed by others
Introduction
High-grade endometrial stromal sarcoma (HGESS) is currently defined as a histologically high-grade monomorphic round and/or spindle cell proliferation of endometrial stromal derivation. This category includes such tumors that harbor YWHAE-NUTM2 fusion [1, 2], ZC3H7B-BCOR fusion [3,4,5,6,6], BCOR internal tandem duplication (ITD) [4, 7,8,9] as well as histologically transformed tumors that harbor low-grade endometrial stromal sarcoma (LGESS)-associated fusions [10]. YWHAE-NUTM2 fusion-positive HGESS consists of a high-grade round cell component that is sometimes associated with a low-grade fibrous or fibromyxoid spindle cell component [1, 2, 11, 12]. HGESS harboring ZC3H7B-BCOR fusion demonstrate high-grade spindle cells embedded in myxoid matrix [3, 6]. HGESS with BCOR ITD have morphologic features of both YWHAE-NUTM2 and ZC3H7B-BCOR fusion-positive tumors [7–9]. These tumors collectively overexpress cyclin D1 [13] and/or BCOR [9] and together with transformed LGESS demonstrate intermediate-grade cytologic atypia and appear to follow an aggressive clinical course [2,3,4, 7, 8, 10, 14]. Rare uterine sarcomas harboring EPC1 and BCORL1 fusions and associated with high-grade histology have been recently reported, but their inclusion among HGESS has not been established [15,16,17].
HGESS share extensive morphologic, immunohistochemical, and genetic overlap with non-gynecologic cancers that harbor YWHAE and BCOR abnormalities. BCOR ITD and YWHAE gene rearrangements have been detected in undifferentiated round cell sarcoma, clear cell sarcoma of the kidney, and central nervous system high-grade neuroepithelial tumor [18,19,20,21,22,23]. BCOR fusions underpin bone or soft tissue high-grade undifferentiated sarcoma and ossifying fibromyxoid tumor [20, 24, 25]. NTRK3 upregulation resulting in pan-Trk overexpression was recently found in soft tissue undifferentiated round cell sarcoma with YWHAE rearrangement, BCOR ITD, and BCOR-CCNB3 fusion, clear cell sarcoma of the kidney, and ossifying fibromyxoid tumor with ZC3H7B-BCOR fusion [26]. Trk inhibition with larotrectinib was recently shown to have marked and durable antitumor activity in solid cancers with NTRK fusion [27, 28]. The therapeutic significance of pan-TRK overexpression in tumors that lack NTRK rearrangement is unknown. To confirm NTRK3 RNA upregulation and identify other potential diagnostic and/or therapeutic markers in HGESS, we examined targeted gene expression profiles and pan-Trk immunohistochemical expression in HGESS with various genotypes.
Materials and methods
Case selection
Thirty-five HGESS with available formalin-fixed paraffin-embedded (FFPE) tissue diagnosed from 2009 to 2019 were retrospectively collected from five institutions: Memorial Sloan Kettering Cancer Center (New York, NY), University of British Columbia (Vancouver, Canada), Lions Gate Hospital (Vancouver, Canada), University of Calgary (Calgary, Canada), and Mount Sinai Hospital (Toronto, Canada). The cohort included primary and metastatic tumors harboring YWHAE fusion (n = 12) with NUTM2A (n = 1), NUTM2B (n = 7), and partner not confirmed (n = 4); BCOR fusion (n = 16) with ZC3H7B (n = 15) and EPC1 (n = 1); BCOR ITD (n = 4); and BCORL1 fusion (n = 3) with JAZF1 (n = 2) and EPC1 (n = 1) partners. Gene fusion status was previously confirmed by targeted RNA sequencing platforms, MSK Solid Fusion custom assay based on the ArcherTM Anchored Multiplex PCR technology (n = 18) and TruSight RNA Fusion Panel (Illumina, San Diego, CA) (n = 7), and/or fluorescence in situ hybridization (FISH) (n = 7). BCOR ITD was previously detected by the MSK Solid Fusion assay (n = 2), TruSight RNA Fusion Panel (n = 1), or genomic PCR (n = 1). Available pathology reports and hematoxylin and eosin and immunohistochemical stained slides were reviewed by gynecologic pathologists (AMB, SC, CHL) for confirmation of diagnosis. Tumors with histologic and genetic features typical of HGESS with YWHAE and BCOR abnormalities were included. Tumors with BCORL1 rearrangement that demonstrate histologic features of HGESS, including nuclear enlargement, prominent nuclei, and mitotic activity of at least 10 mitotic figures per 10 high power fields were also included. Clinical data, including patient demographics, treatment, and follow-up, were extracted from electronic medical records.
Gene expression analysis
RNA expression data of 11 HGESS (7 primary, 4 recurrent) harboring YWHAE-NUTM2B (n = 4), ZC3H7B-BCOR (n = 3), JAZF1-BCORL1 (n = 1), and EPC1-CXorf67 (n = 1) fusions, and BCOR ITD (n = 2), were extracted from the MSK Solid Fusion assay version 3 that detects gene fusions and oncogenic isoforms in selected protein-coding exons of 62 genes, as previously described [29]. Unique RNA reads for gene probes were normalized to the average number of unique RNA reads for four separate housekeeping genes interrogated as part of the assay (CHMP2A, GPI, RAB7A, and VCP). Probe expression values were then standardized, and the average expression of genes included in the panel was calculated. Hierarchical clustering was performed at gene and probe levels after normalization for unsupervised clustering and dimensionality reduction. Differential expression analysis was performed at gene level with comparisons made between HGESS and 48 other uterine sarcomas, including LGESS (n = 9), undifferentiated uterine sarcoma (UUS) (n = 23), and conventional (n = 11) and myxoid (n = 5) leiomyosarcoma (LMS), that were previously subjected to targeted RNA sequencing as part of their diagnostic evaluation. Fusion data were available for all LGESS which harbored JAZF1-SUZ12 (n = 6) and JAZF1-PHF1 (n = 1); in two LGESS, no fusion was detected by the MSK Solid Fusion assay. An RBPMS-NTRK3 fusion-positive uterine sarcoma served as reference control.
Immunohistochemistry
A pan-Trk antibody directed against the preserved domain common to TrkA, TrkB, and TrkC proteins (EPR17341; Abcam, Cambridge, MA) was used to stain five-µm FFPE whole tissue sections of 35 HGESS, as previously described [30]. Staining was assessed in the high-grade, and when present, low-grade components of all cases and recorded as cytoplasmic and/or nuclear. Intensity was assessed as weak (1+), moderate (2+), and strong (3+). Extent was reported as focal (<30%), patchy (30–70%), and diffuse (>70%). A combined score was calculated by the intensity multiplied by percentage of positive tumor cells. An RBPMS-NTRK3 fusion-positive uterine sarcoma served as a positive control. The results of ER, PR, cyclin D1, and/or BCOR immunohistochemical stains previously performed in the diagnostic work up of the tumor samples were recorded based on review of available immunostained slides and extracted from pathology reports when immunostained slides were not available.
Results
HGESS represents a distinctive molecular group by RNA expression analysis
Targeted RNA expression analysis was successful in all cases (n = 59/59, 100%). Hierarchical clustering of the top 25% of differentially expressed probes revealed three molecular groups (Fig. 1A). The first group consisted of eight cases (13%), including the majority of HGESS harboring YWHAE and BCOR abnormalities (n = 7/11, 64%) and a single UUS harboring HMGA-RAD51B fusion (n = 1/23, 4%) and demonstrated high expression of NTRK3, FGFR3, RET, BCOR, GLI1, and PTCH1 and low ESR1 expression. Notably, the majority of HGESS demonstrated higher expression of NTRK3, FGFR3, RET, BCOR, GLI1, and PTCH1 compared to other uterine sarcoma types (p value <0.001 and log-fold difference >2 for all genes) (Fig. 1). Upregulation of these genes was more robust in HGESS harboring BCOR ITD or YWHAE fusion compared to HGESS with BCOR fusion (Fig. 1). Notably, the average level of NTRK3 upregulation was higher in HGESS in comparison to the one NTRK3 fusion-positive uterine sarcoma control. NTRK1, NTRK2, FGFR1, and FGFR2 mRNA expression was not significantly elevated in HGESS.
A Heatmap of uterine sarcoma gene expression profile shows clustering of most HGESS that is separate from LGESS, UUS, and LMS. Two HGESS harboring ZC3H7B-BCOR fusion clustered with LGESS. B The line plot shows that the differences in the expression of NTRK3, PTCH1, GLI1, FGFR3, BCOR, and RET genes set HGESS apart from other uterine sarcoma subtypes. C Scatterplots of normalized expression of NTRK3, PTCH1, and ESR1 demonstrate higher expression of NTRK3 and PTCH1 and lower expression of ESR1 among HGESS compared to other uterine sarcoma subtypes. The outlier HGESS with high ESR1 expression harbored a ZC3H7B-BCOR fusion and had low NTRK3 expression (HGESS-6).
The second group consisted of 20 cases (34%), including HGESS (3/11, 27%), LGESS (n = 8/9, 89%), UUS (n = 5/23, 22%), and LMS (n = 4/16, 25%) and demonstrated high ESR1 expression and low expression of NTRK3, FGFR3, RET, BCOR, GLI1, and PTCH1 (Fig. 1) with the most prominent difference observed when comparing LGESS and HGESS (Fig. 1C). The small subset of HGESS in this group included tumors harboring ZC3H7B-BCOR fusion (n = 2) and JAZF1-BCORL1 (n = 1). This group also consisted of the majority of LGESS, including all tumors harboring JAZF1-SUZ12 (n = 7) and JAZF1-PHF1 (n = 1) fusions as well as a small subset of UUS, one of which demonstrated an ESR1-MAMLD1 fusion. No fusions were detected by the MSK Solid Fusion assay in the remaining LGESS and two UUS as well as the small subset of LMS in this group.
The third group contained 31 cases (53%), including HGESS (n = 1/11, 9%), LGESS (n = 1/9, 11%), UUS (n = 17/23, 74%), and LMS (n = 12/16, 75%), and demonstrated low expression of NTRK3, FGFR3, RET, BCOR, GLI1, and PTCH1 along with low ESR1. FOXO1-ATP7B and FAM110B-PLAG1 fusions were detected two myxoid LMS. No fusions were observed in the remaining LGESS, LMS, and UUS by the MSK Solid Fusion assay.
No NTRK fusions were detected among any cases in all three molecular groups. Given tumor rarity, clinical data were insufficient to identify survival differences between groups.
NTRK3 mRNA upregulation is associated with pan-Trk expression in HGESS regardless of genotype or cyclin D1/BCOR positivity
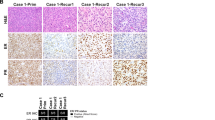
Pan-Trk expression was evaluated in 35 HGESS, including 10 tumors described above in which RNA expression data were available, and an additional 25 tumors with genotypes confirmed by FISH and/or targeted RNA sequencing by MSK Solid Fusion or Trusight RNA Fusion assays. Pan-Trk staining was observed in the vast majority of HGESS (n = 32/35, 91%), including YWHAE fusion- (n = 11/12, 92%), BCOR fusion- (n = 15/16, 94%), BCOR ITD- (n = 4/4, 100%), and BCORL1 fusion- (n = 2/3, 67%) positive tumors (Fig. 2). Staining was cytoplasmic in 31 tumors and nuclear in one which harbored a ZC3H7B-BCOR fusion (Fig. 2). Staining distribution ranged from 10% to 100% of tumor cells with variable intensity and was present in both high- and low-grade components, the latter observed among YWHAE fusion- and BCOR ITD-positive HGESS (Figs. 2A–D and 3A). Staining was moderate to strong and diffuse in all positive YWHAE-rearranged HGESS (Figs. 2A, B and 3). In contrast, HGESS with BCOR fusion and BCOR ITD demonstrated heterogeneous distribution and intensity of pan-Trk expression (Figs. 2C–F and 3). Staining was weak to moderate and diffuse in nine BCOR fusion-positive tumors, including one HGESS harboring EPC1-BCOR fusion (Figs. 2E, F and 3). Pan-Trk staining was focal with variable intensity in two BCORL1 fusion-positive tumors (Fig. 3). The average combined pan-Trk score was 2.13, 1.62, 1.12, and 0.53 in tumors harboring YWHAE fusion, BCOR ITD, BCOR fusion, and BCORL1 fusion, respectively, compared to 2.40 in the NTRK fusion-positive control (Figs. 2G, H and 3). No staining was seen in three HGESS harboring YWHAE, BCOR, and BCORL1 rearrangements.
The high-grade round cell component of two HGESS harboring (A) YWHAE-NUTM2 fusion and (C) BCOR ITD demonstrate (B, D) strong and diffuse cytoplasmic expression of pan-Trk. E, F Focal and strong nuclear pan-Trk expression is seen in a HGESS harboring ZC3H7B-BCOR fusion. G, H Moderate and diffuse pan-Trk expression in a uterine sarcoma with NTRK3 rearrangement is shown as a comparison.
To compare RNA expression analysis and pan-Trk immunohistochemistry, 14 HGESS in which both techniques were performed were evaluated. Pan-Trk staining scores were compared with NTRK3 mRNA expression levels in all cases and showed good correlation across HGESS genotypes (Pearson’s R: 0.631, p value: 0.005) (Supplementary Fig. 1).
Cyclin D1 and/or BCOR immunohistochemistry was previously performed as part of the diagnostic evaluation in 19 HGESS and at least one of the markers was diffusely positive in 14 tumors. Pan-Trk was expressed with variable intensity and extent of staining in all 14 tumors demonstrating diffuse cyclin D1 and/or BCOR immunoreactivity. Among five HGESS in which cyclin D1 and/or BCOR expression was only focal or patchy, pan-Trk expression was positive in all tumors with variable intensity and extent of staining.
ER and PR expression may be seen in HGESS despite ESR1 downregulation
ESR1 expression levels were available for analysis in 11 HGESS and were downregulated in 82% of tumors. ER and PR status were available in eight tumors. Three of them (HGESS-4, HGESS-8, and HGESS-9) were negative for ER and PR by immunohistochemistry and concordant with ESR1 expression levels despite the presence of a low-grade component in the tumor sample of one case submitted for RNA sequencing (HGESS-8) (Table 1). Six tumors showed either focal (n = 4) or diffuse (n = 2) positivity for ER and PR (Table 1). Two tumors harboring ZC3H7B-BCOR (HGESS-6) and JAZF1-BCORL1 (HGESS-2) fusions both demonstrated ER and PR staining and high ESR1 expression. The four remaining tumors (HGESS-1, HGESS-3, HGESS-7, HGESS-11) harboring YWHAE fusion, BCOR fusion, EPC1-CXorf67 fusion, and BCOR ITD, the latter demonstrating a low-grade component in the tumor sample submitted for RNA sequencing, showed ER and PR staining despite low ESR1 expression. Two patients with ER and PR positive and ESR1 downregulated stage I HGESS harboring YWHAE fusion (HGESS-3) and BCOR ITD (HGESS-7) were treated postoperatively with megestrol acetate and letrozole, respectively. Both patients recurred at 12 and 36 months after primary resection, respectively. Six patients did not receive adjuvant hormonal therapy. No treatment history was available in the remaining three cases which were pathology consultations only. ER and PR status were not known for two HGESS (HGESS-5 and HGESS-10).
Discussion
HGESS represents a morphologically, immunohistochemically, and genetically diverse group of uterine sarcomas. Through unsupervised clustering analysis of RNA expression data extracted from targeted RNA sequencing, we showed that 64% of HGESS regardless of genotype form a distinct molecular group defined by upregulation of NTRK3, RET, FGFR3, PTCH1, and GLI1 along with downregulation of ESR1 expression. Frequent pan-Trk expression with variable intensity and extent was observed in over 90% of HGESS harboring YWHAE, BCOR, and BCORL1 fusions as well as BCOR ITD, correlating with NTRK3 mRNA overexpression in a subset where both modalities were performed. ESR1 downregulation was demonstrated in the majority of HGESS for which RNA expression data were available, despite ER and PR expression in 44% of tumors. Two patients with ER and PR positive and ESR1 downregulated stage I HGESS were treated postoperatively with adjuvant endocrine therapy and recurred 12 and 36 months after primary resection. NTRK overexpression at RNA and protein levels and ESR1 downregulation may provide clinical and therapeutic implications for patients with HGESS.
For the first time, we demonstrated high level expression of NTRK3, RET, and FGFR3 kinases and upregulation of sonic hedgehog (SHH) pathway genes, PTCH1 and GLI1, in most HGESS, distinguishing them from other uterine sarcoma subtypes such as LGESS, LMS, and UUS. These genes are associated with neuronal reprogramming in neural crest [31] with reactivation of neuronal reprogramming previously established in various cancers [32, 33]. In HGESS, it has been shown that tumors harboring YWHAE fusion express neuroepithelial markers such as CD56 and CD99 [34]. However, it remains to be seen whether the expression profile observed in HGESS represents direct neuronal or a more primitive stem cell conversion. Interestingly, our findings contrast with those reported in a separate study in which by microarray analysis, GLI1 and RET expression was downregulated in seven HGESS harboring YWHAE-NUTM2 and other rare novel fusions compared to LGESS harboring JAZF1-SUZ12 fusion [35]. Additionally, NTRK3, FGFR3, PTCH1, and ESR1 were not differentially expressed among HGESS and LGESS in that study [35]. Discordance between these findings may be due to the different technologies utilized and the number of genes assessed.
As expected from RNA expression data, the vast majority of HGESS demonstrated predominately cytoplasmic and rarely nuclear pan-Trk expression by immunohistochemistry. Since there was good correlation between NTRK3 mRNA and pan-Trk expression and no evidence of NTRK1 or NTRK2 overexpression, positive pan-Trk staining was likely secondary to NTRK3 upregulation in HGESS. While the extent and intensity of staining were variable in HGESS, diffuse and moderate/strong staining was seen in over 50% of positive tumors. Interestingly, pan-Trk staining was present in the low-grade component of tumors with YWHAE fusion or BCOR ITD as well as BCOR fusion-positive tumors displaying only mild nuclear atypia. Pan-Trk expression was also not associated with cyclin D1 or BCOR expression, the latter which may be variable in BCOR fusion-positive HGESS.
Pan-Trk immunohistochemistry when used alongside cyclin D1 and BCOR expression in the correct clinical context may serve as a diagnostic marker for HGESS with YWHAE, BCOR, and BCORL1 genetic abnormalities when molecular confirmation is not available. Cyclin D1 has a sensitivity and specificity of 100% and 99%, respectively, for detecting YWHAE fusion-positive HGESS [13]. BCOR has a sensitivity and specificity of 87% and 100%, respectively, for detecting HGESS with YWHAE or BCOR abnormalities [9]. While over 70% of NTRK fusion-positive uterine sarcomas may express diffuse cyclin D1, BCOR expression is negative or rarely seen in <5% of tumor cells [36]. Based on these findings, a diagnosis of HGESS may be considered in any uterine sarcoma demonstrating both pan-Trk of any intensity or extent and diffuse BCOR positivity of any intensity, regardless of cyclin D1 expression status.
Upregulation of NTRK3 expression has been recently demonstrated in a variety of tumor types. Increased NTRK3 expression at protein and RNA levels was recently shown in soft tissue sarcomas harboring YWHAE fusions and BCOR ITD as well as ossifying fibromyxoid tumors with BCOR fusion, all of which share overlapping histologic features with HGESS underpinned by identical genetic aberrations [26]. In addition, desmoplastic small round cell tumor and salivary gland adenoid cystic carcinoma harboring EWSR1-WT1 and MYB-NFIB fusions, respectively, were shown to have increased NTRK3 expression [37], resulting in activation of neural reprograming [32] and TrkC proteins [38, 39]. The upregulation and activation of NTRK mRNA and Trk proteins appears to be specific to NTRK3 and TrkC [26, 39]; this activation may be due to chromatin remodeling and alteration of gene silencing due to BCOR genetic aberrations [40, 41] or other epigenetic alterations, however, the exact mechanism of activation needs to be investigated.
Given the efficacy of larotrectinib and entrectonib in patients with NTRK fusion-positive tumors [42], pan-Trk immunohistochemistry has been proposed as a cost-effective and widely available tool to screen for tumors which may harbor potentially treatable NTRK fusions [39, 43, 44]. However, based on our current and previous studies, pan-Trk staining should not be considered a surrogate marker of an NTRK fusion event. We previously showed that Trk staining may be seen in 3% of uterine LMS lacking NTRK rearrangement and is likely secondary to physiologic expression in smooth muscle [45]. Based on our findings in the current study, pan-Trk expression is seen in over 90% of HGESS with frequency of staining similar to soft tissue counterparts [26]. Furthermore, the overall sensitivity and specificity of pan-Trk immunohistochemistry vary by tumor type and are only 80 and 74%, respectively, in soft tissue sarcomas [39]. These observations suggest that NTRK, BCOR, BCORL1, and YWHAE gene rearrangement as well as BCOR ITD status should be assessed in any uterine sarcoma demonstrating pan-Trk expression of any extent.
Downregulation of ESR1 expression in our cohort of HGESS appears to support the observation of absent ER and PR expression in the high-grade component of YWHAE fusion-positive HGESS and the majority of ZC3H7B-BCOR fusion-positive HGESS. While ESR1 downregulation was observed in most HGESS in our study, a subset of tumors demonstrated focal ER and PR positivity. Interestingly, two patients with stage I ER and PR positive HGESS in our study were treated with endocrine therapy, and both patients recurred after 12 and 36 months postoperatively. Despite ER and PR immunoreactivity, ESR1 expression was downregulated in both tumors. These findings suggest that while ER and PR staining may be seen in some HGESS, the low ESR1 RNA expression may explain the limited efficacy of endocrine therapy in these tumors.
In conclusion, HGESS exhibits morphologic and genetic diversity, but appear homogenous by RNA expression. They are distinct from other uterine sarcoma subtypes by activation of NTRK3, FGFR3, and RET kinases as well as SHH pathway genes, PTCH1 and GLI1, along with downregulation of ESR1. Like soft tissue sarcomas bearing YWHAE and BCOR genetic abnormalities, the majority of HGESS demonstrate pan-Trk staining which may serve as a diagnostic biomarker. ESR1 downregulation is seen in some HGESS that express ER and PR which raises implications in the utility of endocrine therapy in these patients.
References
Croce S, Hostein I, Ribeiro A, Garbay D, Velasco V, Stoeckle E, et al. YWHAE rearrangement identified by FISH and RT-PCR in endometrial stromal sarcomas: genetic and pathological correlations. Mod Pathol. 2013;26:1390–400.
Lee CH, Marino-Enriquez A, Ou W, Zhu M, Ali RH, Chiang S, et al. The clinicopathologic features of YWHAE-FAM22 endometrial stromal sarcomas: a histologically high-grade and clinically aggressive tumor. Am J Surg Pathol. 2012;36:641–53.
Lewis N, Soslow RA, Delair DF, Park KJ, Murali R, Hollmann TJ, et al. ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol. 2018;31:674–84.
Momeni-Boroujeni A, Chiang S. Uterine mesenchymal tumours: recent advances. Histopathol. 2020;76:64–75.
Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, Davidson B, et al. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X; 22-translocation. Genes Chromosomes Cancer. 2013;52:610–8.
Hoang LN, Aneja A, Conlon N, Delair DF, Middha S, Benayed R, et al. Novel high-grade endometrial stromal sarcoma: a morphologic mimicker of myxoid leiomyosarcoma. Am J Surg Pathol. 2017;41:12–24.
Juckett LT, Lin DI, Madison R, Ross JS, Schrock AB, Ali S. A pan-cancer landscape analysis reveals a subset of endometrial stromal and pediatric tumors defined by internal tandem duplications of BCOR. Oncology. 2019;96:101–9.
Marino-Enriquez A, Lauria A, Przybyl J, Ng TL, Kowalewska M, Debiec-Rychter M, et al. BCOR internal tandem duplication in high-grade uterine sarcomas. Am J Surg Pathol. 2018;42:335–41.
Chiang S, Lee CH, Stewart CJR, Oliva E, Hoang LN, Ali RH, et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol. 2017;30:1251–61.
Zou Y, Turashvili G, Soslow RA, Park KJ, Croce S, McCluggage WG, et al. High-grade transformation of low-grade endometrial stromal sarcomas lacking YWHAE and BCOR genetic abnormalities. Mod Pathol. 2020;33:1861–70.
Lee CH, Ou WB, Marino-Enriquez A, Zhu M, Mayeda M, Wang Y, et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci USA. 2012;109:929–34.
Sciallis AP, Bedroske PP, Schoolmeester JK, Sukov WR, Keeney GL, Hodge JC, et al. High-grade endometrial stromal sarcomas: a clinicopathologic study of a group of tumors with heterogenous morphologic and genetic features. Am J Surg Pathol. 2014;38:1161–72.
Lee CH, Ali RH, Rouzbahman M, Marino-Enriquez A, Zhu M, Guo X, et al. Cyclin D1 as a diagnostic immunomarker for endometrial stromal sarcoma with YWHAE-FAM22 rearrangement. Am J Surg Pathol. 2012;36:1562–70.
Parra-Herran C, Howitt BE. Uterine mesenchymal tumors: update on classification, staging, and molecular features. Surg Pathol Clin. 2019;12:363–96.
Dickson BC, Lum A, Swanson D, Bernardini MQ, Colgan TJ, Shaw PA, et al. Novel EPC1 gene fusions in endometrial stromal sarcoma. Genes Chromosomes Cancer. 2018;57:598–603.
Allen AJ, Ali SM, Gowen K, Elvin JA, Pejovic T. A recurrent endometrial stromal sarcoma harbors the novel fusion JAZF1-BCORL1. Gynecol Oncol Rep. 2017;20:51–3.
Muthukumarana V, Fix DJ, Stolnicu S, Park KJ, Soslow RA, Benayed R, et al. BCOR expression in mullerian adenosarcoma: a potential diagnostic pitfall. Am J Surg Pathol. 2020;44:765–70.
Kao YC, Sung YS, Zhang L, Huang SC, Argani P, Chung CT, et al. Recurrent BCOR internal tandem duplication and YWHAE-NUTM2B fusions in soft tissue undifferentiated round cell sarcoma of infancy: overlapping genetic features with clear cell sarcoma of kidney. Am J Surg Pathol. 2016;40:1009–20.
Yoshida Y, Nobusawa S, Nakata S, Nakada M, Arakawa Y, Mineharu Y, et al. CNS high-grade neuroepithelial tumor with BCOR internal tandem duplication: a comparison with its counterparts in the kidney and soft tissue. Brain Pathol. 2018;28:710–20.
Wong MK, Ng CCY, Kuick CH, Aw SJ, Rajasegaran V, Lim JQ, et al. Clear cell sarcomas of the kidney are characterised by BCOR gene abnormalities, including exon 15 internal tandem duplications and BCOR-CCNB3 gene fusion. Histopathol. 2018;72:320–9.
Roy A, Kumar V, Zorman B, Fang E, Haines KM, Doddapaneni H, et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun. 2015;6:8891.
Ueno-Yokohata H, Okita H, Nakasato K, Akimoto S, Hata J, Koshinaga T, et al. Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet. 2015;47:861–3.
Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–72.
Kao YC, Owosho AA, Sung YS, Zhang L, Fujisawa Y, Lee JC, et al. BCOR-CCNB3 fusion positive sarcomas: a clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas. Am J Surg Pathol. 2018;42:604–15.
Antonescu CR, Sung YS, Chen CL, Zhang L, Chen HW, Singer S, et al. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors–molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–93.
Kao YC, Sung YS, Argani P, Swanson D, Alaggio R, Tap W, et al. NTRK3 overexpression in undifferentiated sarcomas with YWHAE and BCOR genetic alterations. Mod Pathol. 2020;33:1341–9.
Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in tRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–9.
Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531–40.
Benayed R, Offin MD, Mullaney KA, Sukhadia P, Rios KM, Desmeules P, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low mutational burden. Clin Cancer Res. 2019;25:4712–22.
Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41:1547–51.
Masserdotti G, Gascón S, Götz M. Direct neuronal reprogramming: learning from and for development. Development. 2016;143:2494–510.
Kang H-J, Park JH, Chen W, Im Kang S, Moroz K, Ladanyi M, et al. EWS–WT1 oncoprotein activates neuronal reprogramming factor ASCL1 and promotes neural differentiation. Cancer Res. 2014;74:4526–35.
Jiang Y-y, Wei M, Zhang H-z. Tumor cells undergoing direct lineage conversion to neurons: unnatural but useful? Neural Regen Res. 2015;10:334.
McCluggage WG, Lee C-H. YWHAE-NUTM2A/B translocated high-grade endometrial stromal sarcoma commonly expresses CD56 and CD99. Int J Gynecol Pathol. 2019;38:528–32.
Micci F, Gorunova L, Agostini A, Johannessen LE, Brunetti M, Davidson B, et al. Cytogenetic and molecular profile of endometrial stromal sarcoma. Genes Chromosomes Cancer. 2016;55:834–46.
Croce S, Hostein I, Longacre TA, Mills AM, Perot G, Devouassoux-Shisheboran M, et al. Uterine and vaginal sarcomas resembling fibrosarcoma: a clinicopathological and molecular analysis of 13 cases showing common NTRK-rearrangements and the description of a COL1A1-PDGFB fusion novel to uterine neoplasms. Mod Pathol. 2019;32:1008–22.
Spraggon L, Hmeljak J, Magnan H, Somwar R, Asher M, Hartono AB, et al. Abstract B12: therapeutic potential of NTRK3 in desmoplastic small round cell tumor [abstract]. In: Proceedings of the AACR conference on advances in sarcomas: from basic science to clinical translation. Philadelphia, PA: AACR; 2017. Clin Cancer Res 2018;24(2_Suppl):Abstract nr B12.
Ivanov S, Panaccione A, Brown B, Guo Y, Moskaluk C, Wick M, et al. TrkC signaling is activated in adenoid cystic carcinoma and requires NT-3 to stimulate invasive behavior. Oncogene. 2013;32:3698–710.
Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020;33:38–46.
Bitoun E, Davies KE. The robotic mouse: understanding the role of AF4, a cofactor of transcriptional elongation and chromatin remodelling, in purkinje cell function. Cerebellum. 2009;8:175–83.
Astolfi A, Fiore M, Melchionda F, Indio V, Bertuccio SN, Pession A. BCOR involvement in cancer. Epigenomics. 2019;11:835–55.
Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731–47.
Solomon JP, Benayed R, Hechtman JF, Ladanyi M. Identifying patients with NTRK fusion cancer. Ann Oncol. 2019;30:viii16–22.
Solomon JP, Hechtman JF. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. 2019;79:3163–8.
Chiang S, Cotzia P, Hyman DM, Drilon A, Tap WD, Zhang L, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol. 2018;42:791–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SY is an advisory board member and received travel allowance from Amgen, AstraZeneca, Bayer, Novartis, and Roche. SC is a consultant for AstraZeneca. All remaining authors have declared no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Momeni-Boroujeni, A., Mohammad, N., Wolber, R. et al. Targeted RNA expression profiling identifies high-grade endometrial stromal sarcoma as a clinically relevant molecular subtype of uterine sarcoma. Mod Pathol 34, 1008–1016 (2021). https://doi.org/10.1038/s41379-020-00705-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-00705-6
This article is cited by
-
Uterine leiomyoma with RAD51B::NUDT3 fusion: a report of 2 cases
Virchows Archiv (2023)
-
Adult NTRK-rearranged spindle cell neoplasms of the viscera: with an emphasis on rare locations and heterologous elements
Modern Pathology (2022)
-
CNS tumors with YWHAE:NUTM2 and KDM2B-fusions present molecular similarities to extra-CNS tumors having BCOR internal tandem duplication or alternative fusions
Acta Neuropathologica Communications (2021)