Abstract
Cellular proliferation is correlated with the progression of melanoma. Accordingly, the proliferation index of H&E-stained thin melanomas was recently included in the staging system of the American Joint Committee on Cancer. Yet, the immunohistochemical markers of proliferation phosphohistone H3 and Ki67 may improve such indices. To accurately quantify these markers, they should be combined with a melanocytic marker, for example, MART1 in an immunohistochemical double stain; also enabling automated quantification by image analysis. The aim of the study was to compare the prognostic impact of phosphohistone H3/MART1, Ki67/MART1, and H&E stains in primary cutaneous melanoma, and to determine the difference between indices established in hot spots and the global tumor areas. The study included 153 consecutive stage I/II melanoma-patients. The follow-up time was 8–14 years for event-free melanoma. Recurrent disease occurred in 43 patients; 37 died of melanoma. Both events occurred in only three thin melanomas. Their paraffin-embedded tissue was stained for phosphohistone H3/MART1, Ki67/MART1, and with H&E. And proliferation indices were established in 1-mm2 hot spots and in the global tumor areas. In multivariate Cox analyses, only hot spot indices of phosphohistone H3/MART1 and Ki67/MART1 were independent prognostic markers. Phosphohistone H3/MART1 tended to be better than Ki67/MART1 with adjusted hazard ratios of 3.66 (95% CI, 1.40–9.55; P=0.008) for progression-free survival and 3.42 (95% CI, 1.29–9.04; P=0.013) for melanoma-specific death. In all stains, prognostic performance was substantially improved by using hot spots instead of the global tumor areas. In conclusion, phosphohistone H3/MART1 and Ki67/MART1 were superior to H&E stains, and hot spots superior to the global tumor areas. Given the potential for automated analysis, these double stains seem to be robust alternatives to conventional mitotic detection by H&E in stage I/II melanomas in general. This was particularly true for thick melanomas whereas no specific analyses for thin melanomas only could be performed.
Similar content being viewed by others
Main
Unlimited cell division is a hallmark of carcinogenesis, and increased cellular proliferation is correlated with declined patient survival in a variety of human malignancies.1, 2, 3 Accordingly in thin primary cutaneous melanoma (≤1 mm), the mitotic index replaced Clark level of invasion in the 2009 staging system of the American Joint Committee on Cancer.4, 5 The recommended quantification of proliferation by hot spots (areas with most mitoses) on H&E stains is, however, widely criticized. Primary concerns are difficulty discerning the mitotic figures, low reproducibility, and excessive time-consumption.6, 7, 8, 9
An alternate option may be immunohistochemical markers of proliferation. The immunohistochemical marker Ki67 labels the nucleus of cells in the active phases of the cell cycle (G1, S, G2, and M)10 and has been extensively studied in both melanoma and other malignancies.8, 11, 12 Most studies demonstrate a correlation between the proportion of Ki67-positive melanocytic cells (Ki67 index) and clinical outcome of melanoma; however, conclusions of studies are contradictory.8
Whereas Ki67 marks nuclei in all active phases of the cell cycle, the proliferation marker phosphohistone H3 (PHH3) visualizes only the four actual phases of mitosis and late G2.13 Recent studies suggest that PHH3 is a more reliable marker of mitosis in melanoma than H&E stains and that time spend to determine the proliferation index may be reduced by at least 50%.14, 15 The prognostic impact of PHH3 is already demonstrated in other malignancies, for instance, breast cancer and meningiomas,16, 17, 18 and recently, similar tendencies were demonstrated in nodular melanomas.19
Yet, all current studies base their analysis of PHH3 and Ki67 on immunohistochemical single stains. Recent studies, however, demonstrate advantages of combining PHH3 and Ki67 with a melanocytic marker, for example, melanoma antigen recognized by T cells (MART1) in an immunohistochemical double stain.15, 20, 21 Thus, because double stains enable clear distinction between proliferative melanocytic cells and proliferative lymphocytes, histiocytes, stromal cells, and endothelial cells, the prognostic impact of Ki67 and PHH3 may be improved. In addition, fast and repeatable quantification by automated image analysis (AIA) is possible.22
Today, Ki67 indices are mostly quantified in the entire tumor section, whereas indices of PHH3 and H&E stains are quantified in hot spots. The difference between these two techniques and their influence on clinical staging are largely unexplored.
The aim of this study was to compare the prognostic impact of proliferation in PHH3/MART1-, Ki67/MART1-, and H&E-stained primary cutaneous melanomas and, additionally, to establish the most favorable reference space for proliferative activity (hot spot vs the global tumor area) in melanoma prognosis.
Materials and methods
Specimens
Formalin-fixed and paraffin-embedded tissue from 190 primary cutaneous melanomas was retrieved from the archives of the Department of Pathology at Aarhus University Hospital, Aarhus, Denmark and Randers Hospital, Randers, Denmark. The prospective patient cohort, previously described in detail,23 included consecutive stage I/II patients from February 1997 to December 2000; an era before the introduction of the sentinel node technique.23
The study was conducted after approval by the Central Denmark Region Committee on Biomedical Research Ethics.
Immunohistochemistry
PHH3/MART1 stains were performed on BenchMark XT (Ventana Medical Systems, Tucson, AZ, USA) by an indirect sequential immunoenzymatic technique. A 3-μm section was cut from each tissue block, mounted on Superfrost Plus slides (Thermo Fisher Scientific, Waltham, MA, USA), and dried for 1 h at 60°C. Standard settings and reagent kits of Benchmark XT (Ventana) were used in deparaffinization, rehydration, antigen retrieval, and endogenous peroxidase blocking. Polyclonal rabbit antibody Phospho-Histone H3 (Ser10, dilution 1:300; Cell Signaling Technology, Danvers, MA, USA) incubated 32 min at room temperature follow by Ventanas ultraView Universal 3,3′-Diaminobenzidin (DAB) Detection Kit. After denaturation (87 °C), monoclonal mouse antibody MART1 (Clone A103, dilution 1:30; Dako Denmark A/S, Glostrup, Denmark) incubated 40 min at room temperature followed by Ventanas ultraView Universal Alkaline Phosphatase Red Detection Kit. Slides were counterstained with Mayer’s hematoxylin and bluing reagent and later manually dehydrated and mounted.
Ki67/MART1 stains were performed on Autostainer Link 48 (Dako) by an indirect simultaneous immunoenzymatic technique. Our Ki67/MART1 procedure has previously been described.20 In short, pretreatment was followed by incubation with monoclonal rabbit antibody Ki67 (Clone SP6; Thermo Fisher Scientific) and monoclonal mouse antibody MART1 (Clone A103; Dako). The visualization system was a polymer mixture of goat anti-rabbit antibody conjugated to horseradish peroxidase [EnVision+ System-HRP (DAB); Dako] and goat anti-mouse antibody conjugated to alkaline phosphatase [Histofine Simple Stain AP (M), Cosmo Bio, Tokyo, Japan]. Liquid Permanent Red (Dako) was applied; then DAB (Dako).20
All PHH3/MART1 and Ki67/MART1 series included positive controls. Ki67/MART1 stains also include internal controls (presence of Ki67/MART1 positivity in epidermis or dermal–epidermal junction).
Quantification of Proliferation Indices
Whole slide images of PHH3/MART1, Ki67/MART1, and H&E stains were captured by Nanozoomer (Hamamatsu Phototonics K.K., Hamamatsu City, Japan) at a magnification of × 20 and saved in their image format NDPI.
A Master of Science (PSN) analyzed the immunohistochemical stains, and an experienced pathologist evaluated H&E slides (TS); both without knowledge of patients’ outcome.
PHH3/MART1 stains
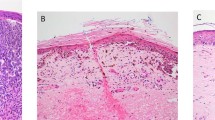
The area with most dermal PHH3/MART1-positive cells was covered by a fixed 1-mm2 square (Figure 1a) in Visiopharm Integrator System 4.2.3.0 [(VIS) Visiopharm A/S, Hoersholm, Denmark]. The number of PHH3/MART1-positive cells manually counted at × 40 in this frame was reported as the PHH3 index in hot spot.
Nodular melanoma stained for PHH3/MART1 (a–d) and Ki67/MART1 (e–h), and with H&E (i, j). (a) A 1-mm2 hot spot. (b) Automated quantification of MART1-verified tumor areas (AMART1, red) within the ROI (green line). (c, d) Positivity of PHH3 and MART1. (e) Automated Ki67 index in hot spot. (f) Automated Ki67 index in the global tumor area. (g) Positivity of Ki67 and MART1. (h) Automated recognition of Ki67-positive (green) and Ki67-negative (blue) melanocytic cells that are verified by MART1 (red). (i) ROI (green line). (j) Automated area-quantification of ROI (AROI, purple).
Dermal PHH3/MART1-positive cells were also counted manually throughout the tumor section. The tumor area was then automatically quantified by the Visiomorph DP module in VIS (Visiopharm). That is, a region of interest (ROI) was manually outlined by the observer (Figure 1b), and the MART1-verified tumor area was quantified by previously described algorithms (Figure 1b).22 The PHH3 index was calculated by

where NPHH3/MART1 is the number of PHH3/MART1-positive cells (Figures 1c and d) and AMART1 the tumor area verified by MART1 (Figure 1b).
Because we occasionally experienced weak intensity of MART1 in PHH3/MART1 stains, the tumor area of such stains (n=11) was merely the area of ROI (Figure 1b).
Ki67/MART1 stains
Visiomorph DP in VIS (Visiopharm) automatically quantified Ki67 indices. The AIA protocol that was fixed and applied to all whole slide images has previously been described.22 Prior to AIA, a hot spot area was manually covered with a 1-mm2 square (Figure 1e), and ROIs were drawn around tumor areas (Figure 1f) in VIS (Visiopharm). The Ki67 index was then automatically calculated for the 1-mm2 hot spot (Figure 1e) and the global tumor area (Figure 1f) by

where NKi67/MART1 is the number of Ki67-positive melanocytic cells and NNeg./MART1 the number of Ki67-negative melanocytic cells (Figures 1g and h).
Hematoxylin and eosin stains
Dermal melanocytic mitoses in H&E stains were manually counted by the conventional hot spot technique24 and in the global tumor areas. Counting was conducted on whole slide images in the Nanozoomer Digital Pathology Viewer (Hamamatsu Phototonics) at × 40.
Again, ROIs were outlined (Figure 1i) in VIS (Visiopharm), and their areas established by AIA (Figure 1j). The number of mitoses in 1 mm2 were reported as the H&E index in hot spot, while the global H&E index was calculated by

where NMitoses is the number mitoses in the global tumor area and AROI the area of tumor (Figure 1j).
Data and Statistical Analyses
Statistical analyses were performed in Stata 10.1 (StataCorp, College Station, TX, USA).
Time to end points, recurrent disease or melanoma-specific death, was measured from the time of diagnosis. Patients who either died without evidence of melanoma or were alive without recurrent disease at last clinical follow-up were censored. Time-to-event data were updated July 2011.
Kaplan–Meier survival curves were based on median indices of PHH3/MART1, Ki67/MART1, and H&E stains. And the groups divided by the median were compared by log-rank tests. Univariate and multivariate analyses were performed using the Cox proportional hazards model.25 Here, patients with missing data were excluded. In multivariate analyses, the directive of minimum 10 events per covariate was followed.26 The model included the recognized prognostic factors, Breslow thickness and ulceration,4 in addition to the relevant proliferation index (positively skewed) that was categorized based on its median. The Cox proportional hazards assumption for all indices and covariates were assessed by log–log plots,27 and linearity of continuous covariates was assessed by the quartile design variable method;28 hence, age at diagnosis was categorized. In addition, Breslow thicknesses were logarithmically transformed.
In all statistical analyses, two-sided P-values <0.05 were considered statistically significant.
Results
Only 153 of the initial 190 patients23 were included in the study. The exclusion criteria were inadequate quality of the tissue section or immunohistochemical stain despite repeated staining attempts (n=7) or insufficient tumor material left for analysis because of excessive sectioning (n=30, the initial median Breslow thickness of this group was 0.6 mm).
Characteristics of the included study patients are presented in Table 1. Of the 153 patients, 43 (28%) experienced recurrent disease and 49 (32%) died; 37 (24%) deaths were related to melanoma. For patients with thin melanomas, only three experienced recurrent disease, and subsequently they all died of melanoma. The median follow-up time was 12 years (range, 8–14 years) for patients with event-free melanoma.
In AIA of Ki67/MART1 stains, one outlier with a very high Ki67 index was excluded from further analysis. The automated indices of this lesion were based on the recognition of only two melanocytic cells.
Kaplan–Meier plots for proliferation indices of PHH3/MART1, Ki67/MART1, and H&E stains are shown in Figure 2 for progression-free survival and in Figure 3 for melanoma-specific death. The divisions of patients in the Kaplan–Meier plots are compared in Table 2. The univariate and final multivariate Cox regression analyses are shown in Tables 3 and 4, respectively. In the multivariate analysis of PHH3 in hot spots, the adjusted hazard ratio (HR) of the Breslow thickness (continuous, log-transformed) was 2.91 (95% CI, 1.60–5.27; P<0.001) for progression-free survival and 3.36 (95% CI, 1.81–6.27; P<0.001) for melanoma-specific death. For ulceration, the adjusted HR was 1.25 (95% CI, 0.574–2.74; P=0.571) for progression-free survival and 1.03 (95% CI, 0.441–2.39; P=0.953) for melanoma-specific death. Similar changes in the adjusted ratios of the Breslow thickness and ulceration were seen in all proliferation indices (data not shown).
Discussion
Increased cellular proliferation is correlated with declined patient survival in a variety of human malignancies.2, 3, 4 Correspondingly, the mitotic index was recently included in the staging system of the American Joint Committee on Cancer in thin melanoma.4, 5 The recommended index-quantification on H&E stains is, however, widely criticized because of presumed inaccuracy, low reproducibility, and excessive time-consumption.6, 7, 8, 9 An alternate option may be immunohistochemical markers; such as PHH3 or Ki67. To accurately quantify PHH3 and Ki67, they should be combined with a melanocytic marker in an immunohistochemical double stain;15, 20, 21 to our knowledge, a shortage in all existing investigations. Double stains may, in addition, enable quantification by AIA. In this study we have compared the prognostic impact of proliferation in PHH3/MART1-, Ki67/MART1-, and H&E-stained primary cutaneous melanomas and compared indices of hot spots and the global tumor areas. In this context, we have demonstrated independent prognostic capabilities of PHH3 and Ki67 as opposed to H&E in our consecutive cohort with stage I/II melanomas.
We found proliferation indices of immunohistochemistry to be highly superior to indices of H&E stains. In immunohistochemical stains, PHH3 tended to be better than Ki67. And when comparing reference spaces, the prognostic performance of indices was substantially improved by using hot spots instead of the global tumor areas.
In univariate analyses, strong association between proliferation and clinical outcome was demonstrated (Figures 2 and 3; Tables 2 and 3); except in global H&E indices. Based on HRs of the univariate analyses, PHH3 indices in hot spots were highly superior to other indices and standard risk factors of melanoma (Table 3). And compared with H&E stains, the HR of proliferation increased twofold by using PHH3 in hot spots (Table 3). Indices of Ki67 stains were also better than the comparable indices of H&E stains (Tables 2 and 3).
In multivariate analyses, the only independent prognostic markers of melanoma were PHH3 and Ki67 in hot spots. Both were superior to ulceration, which lost its usual independent prognostic properties. Indices of H&E stains were, on the other hand, highly dependent on Breslow thickness (Table 4).
When comparing the hot spot technique with indices of the global tumor area, noteworthy differences were recognized. In both univariate and multivariate analysis, the difference between methodologies was apparent for all stains (Figures 2 and 3; Tables 2, 3, 4). The prognostic impact of all stains was markedly improved by using a hot spot instead of the global tumor area as reference space. This may indicate that subclones of progressive tumor cells impact clinical outcome of melanoma to a larger extent than the general proliferative activity of the tumor. We find the evidence for this hypothesis convincing because all three stains, PHH3/MART1, Ki67/MART1, and H&E, show this same pattern of variation.
Previously, only two studies performed by the same group have explored the prognostic impact of PHH3 in cutaneous melanomas.19, 29 Most recently, like us, they found that PHH3 in hot spots is an independent prognostic marker as opposed to H&E mitotic indices; however, with a smaller HR than ours. The two studies are, however, not completely comparable because they only evaluated nodular melanomas, and the Breslow thicknesses were subdivided into three groups.19 In their first study, no correlation between PHH3 and clinical outcome was demonstrated. This study was, however, conducted on tissue microarrays,29 which may be problematic when evaluating hot spots.
Our Ki67 results for global indices are in line with many other studies that demonstrate correlation between increased Ki67 expression and declined survival; though, dependent on Breslow thickness.8 Nevertheless, just as many studies designate Ki67 an independent prognostic marker.8 Seemingly, only one study has used the hot spot technique. They also find that Ki67 in hot spots is an independent prognostic marker.30 Possibly, the prognostic impact of other studies could have been improved if the hot spot technique had been used. Additional controversy between studies may occur because a high Ki67 index may simply reflect a lengthened cell cycle or cell-cycle arrest of Ki67-positive cells before they enter actual mitosis.8 To circumvent this problem, PHH3 can be used because PHH3 only marks actual dividing cells in the short, stable mitotic phase;31, 32 producing more reliable results.
In contrast to our study, many studies report independent prognostic significance of H&E mitotic indices; however, these studies often include a very large patient cohort;8 enabling them to detect smaller differences between groups. This prognostic significance of H&E mitotic indices has been strongest in patients with thin melanomas.4 Our study was, however, based on a smaller cohort of patients, and thus only three events occurred in patients with thin melanomas. Therefore, we were unable to make any specific conclusions about this subgroup of patients. To establish specific prognostic properties of PHH3 and Ki67 in thin melanomas, dedicated studies for this purpose are needed. When performing an independent statistical analysis on the subgroup with thick melanomas (>1 mm, data not shown), we found no significant differences from the results presented for the whole group; hence, the prognostic properties of PHH3 and Ki67 did also apply to only thick melanomas.
When counting proliferative cells, most former studies use a subset of smaller coherent frames to evaluate hot spots. We thus speculated whether our 1-mm2 inflexible hot spot was suitable; especially for thin melanomas. Consequently, we also examined the use of a counting frame assembled from four small 0.25-mm2 frames placed in coherent hot spots. The results of our univariate and multivariate analysis for this technique were similar, though, inferior to the use of one big 1-mm2 hot spot (unpublished observations).
In contrast to single marker studies, immunohistochemical markers of proliferation are often superior to H&E mitotic indices in comparative studies on the same patient cohort.19, 29, 33 Like other investigators,7, 19, 34 we believe that quantification errors are considerably more frequent in indices of H&E than PHH3/MART1 and Ki67/MART1 stains. Apoptotic cells and technical artifacts may be difficult to distinguish from mitoses, and the prophase that comprises a large fraction of the entire mitotic phase19 is undetected; hence, mitotic indices of H&E stains may loose their prognostic impact.
An advantage of our study compared with other studies of PHH3 and Ki67 is the use of immunohistochemical double stains that increase the markers’ specificity in tumor-cell evaluation. This is also a likely explanation for our markedly lower index medians compared with former studies.19, 29, 33 For instance, the median PHH3 index in the Ladstein study of nodular melanomas (median Breslow thickness of 3.6 mm) was 28.7 per mm2 (range, 0–148 per mm2)19 compared with our median of 6 per mm2 (range, 0–43 per mm2); also for nodular melanomas (median Breslow thickness of 2.55 mm).
Although not fully validated, we used AIA for Ki67/MART1 quantification. To justify this, we have quantified all Ki67 indices manually and compared manual and automated Ki67 indices. Indices of AIA tended to be somewhat higher than manual estimates, but their ability to foresee clinical outcome was equal (unpublished observations). We have previously seen this tendency for higher indices in AIA in a study aiming to distinguish melanomas from benign nevi.22
In PHH3, we also used AIA to estimate tumor areas. Usually, we observe no difficulties in such quantification. Unfortunately in this study, the MART1 intensity of our PHH3/MART1 stains varied from very weak to very intense. The tumor area verified by MART1 (Figure 1b) was thus difficult to quantify in weak MART1 stains (n=11). The area of ROI, which was used instead, is an appropriate substitute; though, somewhat less accurate (Figure 1b). The varying MART1 intensity also meant that the number of PHH3/MART1-positive cells was difficult to quantify by AIA. Thus, PHH3/MART1-positive cells were only counted manually in this study. But we believe that an optimization of our PHH3/MART1 stains will enable an automated quantification of the PHH3 index. However, the immediate need for automated PHH3-index quantification seems smaller than with Ki67 indices because of the relative low number of counting points. In general, all IHC markers as well as the digital image analysis should be optimized, and, if possible, standardized before utilized in large studies, and especially if applied in routine practice.
In conclusion, cellular proliferation was a strong prognostic marker in our consecutive cohort of patients with stage I/II primary cutaneous melanoma. The quantification method was, however, very important for the prognostic value of the individual markers. Proliferation indices of PHH3/MART1 and Ki67/MART1 stains were highly superior to indices of H&E stains, and hot spots were superior to indices of the global tumor areas in all stains; hence, only PHH3/MART1 and Ki67/MART1 positivity in hot spots were independent prognostic markers of both progression-free survival and melanoma-specific death. This was true for the complete cohort and thick melanomas whereas exclusive analyses for thin melanomas are still needed. Given the prognostic impact of tumor-specific proliferation and the potential for automated analysis, PHH3/MART1 and Ki67/MART stains seem to be robust alternatives to conventional mitotic detection by H&E stains in stage I/II melanomas.
References
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011;144:646–674.
Alkushi A, Abdul-Rahman ZH, Lim P et al. Description of a novel system for grading of endometrial carcinoma and comparison with existing grading systems. Am J Surg Pathol 2005;29:295–304.
van Diest PJ, van der Wall E, Baak JP . Prognostic value of proliferation in invasive breast cancer: a review. J Clin Pathol 2004;57:675–681.
Balch CM, Gershenwald JE, Soong SJ et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 2009;27:6199–6206.
Piris A, Mihm MC, Duncan LM . AJCC melanoma staging update: impact on dermatopathology practice and patient management. J Cutan Pathol 2011;38:394–400.
Weyers W . The magic of numbers: malignant melanoma between science and pseudoscience. Am J Dermatopathol 2011;33:371–378.
Casper DJ, Ross KI, Messina JL et al. Use of anti-phosphohistone H3 immunohistochemistry to determine mitotic rate in thin melanoma. Am J Dermatopathol 2010;32:650–654.
Vereecken P, Laporte M, Heenen M . Significance of cell kinetic parameters in the prognosis of malignant melanoma: a review. J Cutan Pathol 2007;34:139–145.
Schimming TT, Grabellus F, Roner M et al. pHH3 immunostaining improves interobserver agreement of mitotic index in thin melanomas. Am J Dermatopathol 2012;34:266–269.
Scholzen T, Gerdes J . The ki-67 protein: from the known and the unknown. J Cell Physiol 2000;182:311–322.
de Azambuja E, Cardoso F, de Castro G et al. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer 2007;96:1504–1513.
Martin B, Paesmans M, Mascaux C et al. Ki-67 expression and patients survival in lung cancer: systematic review of the literature with meta-analysis. Br J Cancer 2004;91:2018–2025.
Hendzel MJ, Wei Y, Mancini MA et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997;106:348–360.
Tapia C, Kutzner H, Mentzel T et al. Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am J Surg Pathol 2006;30:83–89.
Ikenberg K, Pfaltz M, Rakozy C et al. Immunohistochemical dual staining as an adjunct in assessment of mitotic activity in melanoma. J Cutan Pathol 2012;39:324–330.
Skaland I, Janssen EA, Gudlaugsson E et al. Phosphohistone H3 expression has much stronger prognostic value than classical prognosticators in invasive lymph node-negative breast cancer patients less than 55 years of age. Mod Pathol 2007;20:1307–1315.
Kim YJ, Ketter R, Steudel WI et al. Prognostic significance of the mitotic index using the mitosis marker anti-phosphohistone H3 in meningiomas. Am J Clin Pathol 2007;128:118–125.
Lende TH, Janssen EA, Gudlaugsson E et al. In patients younger than age 55 years with lymph node-negative breast cancer, proliferation by mitotic activity index is prognostically superior to adjuvant!. J Clin Oncol 2011;29:852–858.
Ladstein RG, Bachmann IM, Straume O et al. Prognostic importance of the mitotic marker phosphohistone H3 in cutaneous nodular melanoma. J Invest Dermatol 2012;132:1247–1252.
Nielsen PS, Riber-Hansen R, Steiniche T . Immunohistochemical double stains against Ki67/MART1 and HMB45/MITF: promising diagnostic tools in melanocytic lesions. Am J Dermatopathol 2011;33:361–370.
Puri PK, Valdes CL, Burchette JL et al. Accurate identification of proliferative index in melanocytic neoplasms with Melan-A/Ki-67 double stain. J Cutan Pathol 2010;37:1010–1012.
Nielsen PS, Riber-Hansen R, Raundahl J et al. Automated quantification of MART1-verified Ki67 indices by digital image analysis in melanocytic lesions. Arch Pathol Lab Med 2012;136:627–634.
Jensen TO, Schmidt H, Moller HJ et al. Macrophage markers in serum and tumor have prognostic impact in American Joint Committee on Cancer stage I/II melanoma. J Clin Oncol 2009;27:3330–3337.
Frishberg DP . Protocol for the examination of specimens from patients with melanoma of the skin. Arch Pathol Lab Med 2009;133:1560–1567.
Altman DG . Practical Statistics for Medical Research. Chapman & Hall: London, 1991, pp 367–395.
Peduzzi P, Concato J, Kemper E et al. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol 1996;49:1373–1379.
Kleinbaum DG . Survival analysis: a self-learning text. Springer: New York, 1996, pp 135–144.
Hosmer DW . Applied survival analysis, regression modeling of time-to-event data 2nd ed. John Wiley & Sons, Inc.: Hoboken, 2008, pp 136.
Ladstein RG, Bachmann IM, Straume O et al. Ki-67 expression is superior to mitotic count and novel proliferation markers PHH3, MCM4 and mitosin as a prognostic factor in thick cutaneous melanoma. BMC Cancer 2010;10:140.
Straume O, Sviland L, Akslen LA . Loss of nuclear p16 protein expression correlates with increased tumor cell proliferation (Ki-67) and poor prognosis in patients with vertical growth phase melanoma. Clin Cancer Res 2000;6:1845–1853.
Aoki Y, Moore GE . Comparative study of mitotic stages of cells derived from human peripheral blood. Exp Cell Res 1970;59:259–266.
Pierard GE . Cell proliferation in cutaneous malignant melanoma: relationship with neoplastic progression. ISRN Dermatol 2012;2012:828146.
Henrique R, Azevedo R, Bento MJ et al. Prognostic value of Ki-67 expression in localized cutaneous malignant melanoma. J Am Acad Dermatol 2000;43:991–1000.
Angi M, Damato B, Kalirai H et al. Immunohistochemical assessment of mitotic count in uveal melanoma. Acta Ophthalmol 2011;89:e155–e160.
Acknowledgements
We thank Allan Thorsteinsson, Martin Nielsen, Lone Nielsen, and Helle Johnsen from the Department of Pathology, Vejle Hospital, Denmark, for performing the Ki67/MART1 double stains and Jeanette Baehr Georgsen from the Department of Pathology, Aarhus University Hospital, Denmark, for performing the PHH3/MART1 double stains. In addition, we thank staff at Visiopharm A/S, Hoersholm, Denmark, for technical assistance in AIA. This study was funded by Arvid Nilsson’s Foundation, Denmark, and the Beckett Foundation, Denmark.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Nielsen, P., Riber-Hansen, R., Jensen, T. et al. Proliferation indices of phosphohistone H3 and Ki67: strong prognostic markers in a consecutive cohort with stage I/II melanoma. Mod Pathol 26, 404–413 (2013). https://doi.org/10.1038/modpathol.2012.188
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.188
Keywords
This article is cited by
-
A nomogram incorporating Ki67 to predict survival of acral melanoma
Journal of Cancer Research and Clinical Oncology (2023)
-
Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy
Clinical Epigenetics (2020)
-
POU4F1 promotes the resistance of melanoma to BRAF inhibitors through MEK/ERK pathway activation and MITF up-regulation
Cell Death & Disease (2020)
-
Evaluation of Kindlin-1 and Ki-67 immunohistochemical expression in primary cutaneous malignant melanoma: a clinical series
Applied Cancer Research (2019)
-
Association of Gpx1 fluctuation in cell cycle progression
In Vitro Cellular & Developmental Biology - Animal (2019)