Abstract
It is uncertain whether uterine leiomyosarcoma arises de novo or in preexisting leiomyoma. Leiomyoma-like areas can be seen associated with uterine leiomyosarcoma, raising the possibility of precursor lesions for uterine leiomyosarcoma. In this study, we examined cases of uterine leiomyosarcoma associated with leiomyoma-like areas at the histological, immunohistochemical and DNA level to further evaluate if benign-looking leiomyoma-like and uterine leiomyosarcoma areas are related. Cases of uterine leiomyosarcoma observed at the New York University Medical Center from 1994 to 2007 were reviewed for the presence of leiomyoma-like areas. Of the 26 cases of uterine leiomyosarcoma observed during this period, 18 cases had an associated leiomyoma-like area (five cellular leiomyoma, four symplastic leiomyoma, four cellular and symplastic leiomyoma and five usual type leiomyoma). Sixteen of the 18 cases were examined immunohistochemically for Ki-67, for estrogen receptor, progesterone receptor and for p53. Immunohistochemical profiles were as expected for leiomyoma-like (the mean expression of p53, ER, PR and Ki-67 at 0.3, 63, 75 and 0.6%, respectively), symplastic leiomyoma-like areas (the mean expression of p53, ER, PR and Ki-67 at 0.6, 85, 89 and 5.5%, respectively) and uterine leiomyosarcoma areas (the mean expression of p53, ER, PR and Ki-67 at 52, 38, 39 and 61%, respectively). In six cases, the leiomyoma-like and uterine leiomyosarcoma areas from each case were examined using high-density oligonucleotide array-CGH to determine genetic aberrations in the two areas. Nearly all the genetic aberrations found in leiomyoma-like areas were also found in the corresponding uterine leiomyosarcoma areas. In addition, uterine leiomyosarcoma areas had additional genetic aberrations. The immunohistochemical profiles and genetic aberrations of the examined cases suggest that uterine leiomyosarcoma could arise from the preexisting leiomyoma-like areas that often have a symplastic or cellular morphology.
Similar content being viewed by others
Main
Although many advances have been made in understanding the pathogenesis of carcinomas at various sites, including the uterus, very little is known with regard to the pathogenesis of uterine leiomyosarcoma. Leiomyosarcoma is the most common uterine sarcoma. It comprises approximately 1% of all uterine malignancies. These are aggressive tumors that may be a diagnostic challenge at times.1, 2, 3, 4, 5, 6, 7 We have previously noted the origin of leiomyosarcoma within a leiomyoma,8 as well as the presence of leiomyoma-like areas associated with some uterine leiomyosarcomas.9 In this study, we examined cases of uterine leiomyosarcoma associated with leiomyoma-like areas at the histological, immunohistochemical and DNA level to further evaluate if benign-looking leiomyoma-like and uterine leiomyosarcoma areas are related.
Materials and methods
Cases of uterine leiomyosarcoma observed at the New York University Medical Center from 1994 to 2007 were reviewed for the presence of leiomyoma-like areas on microscopic examination. These leiomyoma-like areas had to be present in continuity with uterine leiomyosarcoma to qualify. Separate leiomyomas were present in many cases, but these were not considered leiomyoma-like areas for the purposes of this study. On microscopic examination, the leiomyoma-like areas and uterine leiomyosarcoma areas merged with each other. Leiomyoma areas were defined as those areas that showed nuclear atypia of only 1+ to 2+ (mild to moderate), except for symplastic leiomyoma areas that showed very large bizarre nuclei, with <5 mitoses/10 HPF. No coagulative tumor cell necrosis was observed. Leiomyosarcomas were defined as showing 2+ to 3+ (moderate to marked) nuclear atypia, with 10 or more mitoses/10 HPF. Coagulative tumor cell necrosis was present in all cases.
Of the total 26 cases of uterine leiomyosarcoma found during this period, 18 had associated leiomyoma-like areas. Sixteen of these 18 cases were examined immunohistochemically with appropriate controls. Briefly, 5-μm tissue sections on adhesive-coated slides were immunostained by mouse monoclonal antibodies for Ki-67 (MIB-1, Immunotech), p53 (Ventana), estrogen receptor (ER, Ventana) and progesterone receptor (PR, Ventana) using Ventana ES immunostainer. The expression of immunohistochemical markers within the tumor was subjectively assessed to the nearest 5%.
DNA Isolation
The DNA was isolated from leiomyoma-like and uterine leiomyosarcoma areas in six of the 18 cases with leiomyoma-like areas. Five- to 10-μm slices from paraffin-embedded tissue block were washed with HemoDee, HemoDee:Ethanol (1:1) and Ethanol (100%) twice each. The MagnaCompact system (Roche Diagnostics Indianapolis, IN) was used to isolate DNA according to the manufacturer's instructions after proteinase K treatment.
CGH Assay
The leiomyoma-like and uterine leiomyosarcoma areas from six cases were examined using high-density oligonucleotide array-CGH (comparative genomic hybridization) technology10 (Agilent 44K arrays) to compare genetic aberrations in these two areas.
Agilent's Human Genome CGH Microarray Kit 44B is a customized, high-definition microarray that was designed using 44 000 probes comprised of 60-mer oligonucleotides designed by Agilent and printed using Agilent Sureprint technology. We designed the array with E-array software 4.0 (Agilent, Palo Alto, CA), every third probe was removed and replaced by probes recognizing the subtelomeric region (1 Mb proximal to telomeres) for 41 subtelomeric regions. The average resolution of the array is 5 kb in subtelomeres and 125 kb in the remaining human genome software.
Test and reference DNAs are digested with Alu I and Rsa I (Promega), and purified with the QIAprep Spin Miniprep kit (Qiagen). Test DNA and reference DNA were labeled with either Cy3-dUTP or Cy5-dUTP (Perkin Elmer) using the Bioprime Array CGH Genomic Labeling kit (Invitrogen). After the labeling reaction, individually labeled test and reference samples were combined and concentrated using Microcon YM-30 filters (Millipore). The hybridization mixture contained the labeled DNAs, 2 × Hybridization buffer (Agilent, Palo Alto, CA), 10 × blocking agent (Agilent,) and Human Cot-1 DNA (Invitrogen). Microarrays were hybridized in an Agilent SureHyb chamber in a rotisserie oven for 24 h at 65°C. Four washing steps were carried out: room temperature with Agilent's Oligo CGH Wash buffer 1 for 5 min, a 37°C wash with Agilent Oligo CGH Wash buffer 2 for 1 min, an acetonitrile rinse at room temperature for 1 min and a 30 s wash in Agilent's Stabilization and Drying Solution at room temperature. All slides were scanned by Agilent Axon 4000B scanner, using the Genepix 4.0 software. Data were obtained by Agilent Feature extraction software 8.0, and then imported into Agilent CGH analytics 3.4 software for analysis.
DNA copy number changes were detected by CGH analytics software 3.4 (Agilent). The statistical algorithm was ADM-1, sensitivity threshold was 6.0 and the moving average window was 1 Mb. To determine that there is a copy number change in a particular locus, three criteria were required to be met. These were positive call by the software, presence of 10 consecutive probes pointing out the same direction and a 1.5-fold average fold difference in the test DNA compared with the reference DNA.
Results
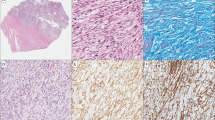
Leiomyoma-like areas were observed to be associated with uterine leiomyosarcoma in 18 of the 26 cases. Of the 18 cases with leiomyoma-like areas, in 5 cases, the morphology of the leiomyoma-like areas was that of a cellular leiomyoma, in 4 cases, the morphology was that of a symplastic leiomyoma (Figure 1a and b), in 4 cases, the morphology was that of a cellular symplastic leiomyoma and in 5 cases, the morphology was that of usual leiomyoma. In all 18 cases, leiomyoma-like and leiomyosarcoma areas merged into each other (Figure 2).
Immunohistochemical profiles were as expected for leiomyoma-like (the mean expression of p53, ER, PR and Ki-67 at 0.3, 63, 75 and 0.6%, respectively), symplastic leiomyoma-like areas (the mean expression of p53, ER, PR and Ki-67 at 0.6, 85, 89 and 5.5%, respectively) and leiomyosarcoma areas (the mean expression of p53, ER, PR and Ki-67 at 52, 38, 39 and 61%, respectively).
Of the six cases examined using array-CGH, three cases were associated with cellular leiomyomas, two cases were associated with symplastic leiomyoma and in one case, the benign component was cellular as well as symplastic.
Table 1 shows the genetic changes observed in uterine leiomyosarcoma and associated leiomyoma-like areas from all six cases.
Chromosomal Changes in Leiomyoma-like Areas
There were more chromosomal losses than gains in leiomyoma-like areas associated with uterine leiomyosarcoma. On an average, there were 2.3 chromosomal gains and 6.5 chromosomal losses in such areas. No chromosomal gains were observed in two cases of leiomyoma-like areas. The average foci of chromosomal gain were five in symplastic leiomyoma-like areas, but only 1.33 in cellular leiomyoma-like areas. Both symplastic and cellular leiomyoma-like areas had a similar level of chromosomal losses. The smallest number of chromosomal changes was observed in case 6, with two gains and one loss, but notably, the entire chromosome 2 was gained. Some of the aberrations observed are listed below. The number in parenthesis indicates the number of cases with this change, followed by significant genes known to be present in those areas.
Chromosomal gains in leiomyoma-like areas and genes located in those areas are as follows:
- • 1p21.3–p31.3 (2/6):
-
JUN
- • 1q22–q24.3 (2/6):
-
CSk1
- • 6q16.3 (1/6):
-
FYN
- • 12p11.22–p13.3 (2/6):
-
K-Ras
- • 12q21.2–q24.3 (2/6):
-
ELK3
Chromosomal losses in leiomyoma-like areas, and genes located in those areas are as follows:
• 2q36.1–q37.2 (2/6)
- • 9p21.3 (2/6):
-
p16, p14ARF
- • 9q33.1–34.2 (2/6):
-
Endoglin
• 16q12.2–24.2 (2/6)
- • 17p13.2 (1/6):
-
p53
- • 22q13.1–22q13.3 (4/6):
-
PDGFB
Chromosomal Changes in Uterine Leiomyosarcoma
Nearly all the chromosomal changes observed in associated leiomyoma-like areas were also present in the leiomyosarcoma in each of the cases (Table 1).
Additional chromosomal losses were also more frequent than were additional chromosomal gains. There were an average of 3.3 additional foci of chromosomal gain and 5 additional foci of chromosomal losses in uterine leiomyosarcoma areas. The minimal number of such additional changes was one gain and three losses, also observed in case 6, but notably, a whole chromosome 4 was lost. Some of the genetic aberrations observed are listed below.
The additional chromosomal gains observed in uterine leiomyosarcoma and associated genes are the following:
- • 8q21.2–q24.23 (2/6):
-
MYC
- • Xp11.3 (2/6):
-
miRNA-221
The additional chromosomal losses observed in uterine leiomyosarcoma and associated genes are the following:
- • 4q34.3–q35.2 (3/6):
-
ING2
- • 8p21.1–p23.2 (2/6):
-
LOXL2
• 11p14.3–p15 (2/6)
- • 13q14.2 (2/6):
-
RB-1
- • 14q13.12–q13.43 (3/6):
-
TTF-1
- • 17p13.2 (1/6):
-
p53
Discussion
The pathogenesis of uterine leiomyosarcoma is poorly understood. It is generally believed that uterine leiomyosarcomas arise de novo, rather than from any precursor lesions. However, cases of uterine leiomyosarcoma arising in leiomyoma have been reported, suggesting that leiomyosarcomas may arise from preexisting leiomyomas.2, 11, 12, 13
In this study, we found benign-looking leiomyoma-like areas in 18 of the 26 cases of uterine leiomyosarcoma. The immunohistochemical profile of these benign-looking areas in this study was similar to that reported for leiomyomas previously.14 The immunohistochemical profile of the leiomyosarcoma areas was similar to that for leiomyosarcomas reported previously.14
We found chromosomal losses and gains in leiomyoma-like areas, with a retention of these losses and gains in the associated leiomyosarcomas. In addition, leiomyosarcomas showed further additional losses and gains of chromosomal areas (Table 1).
Leiomyoma-like areas showed amplification in many oncogene and transcription factor genes, including C-JUN, Cks-1, Fyn, K-Ras and ELK-3. C-JUN forms AP-1 early response transcription factor in association with c-Fos.15 Cks1, located at 1q21, has been proposed recently as an oncogene for breast cancer.16 Fyn is a tyrosine-specific phospho-transferase and activates Ras.17 K-Ras is mutated in many carcinomas. The K-Ras protein is a GTPase and acts as a molecular on/off switch in many signal-transduction pathways.18, 19 ELK-3 activates transcription in the presence of Ras.20
Losses were observed generally in foci with tumor suppressor genes. p16 and p14ARF are tumor suppressors located on 9p21.21 Endoglin has been identified as a candidate tumor suppressor gene in esophageal squamous cell carcinoma.22 PDGFB is a mitogenic factor for mesenchymal cells,23 and its loss is difficult to explain in leiomyoma-like areas. Perhaps other tumor suppressor genes are located in the vicinity of PDGFB, which is at 22q13.1. Deletion of 22q13 is common in ovarian, breast and colorectal cancer.24, 25
Overall, chromosomal losses were more frequent than were chromosomal gains in the leiomyosarcoma area, similar to that which has been reported previously in uterine leiomyosarcomas.26
The amplification of additional chromosomal foci, including that for Myc, was observed in leiomyosarcoma areas. The Myc gene encodes for a transcription factor that regulates the expression of approximately 15% of all genes and has an important function in many malignancies.27 Overexpression of miRNA-221 has been observed in high-grade glioma.28
Additional chromosomal foci, many involving tumor suppressor genes, were lost in leiomyosarcomas. These included foci for ING2, LOXL2, TTF-1, RB-1 and p53. ING2 deletions have been observed in head and neck cancer.29 LOXL2 is frequently lost in prostate carcinoma.30 TTF-1 is a lineage-associated marker for thyroid and lung tissue. It is associated with improved survival, suggesting a potential tumor-suppressing function.31 RB-1 is a tumor suppressor gene that is lost or mutated in many malignancies. Loss of chromosome 13, in which RB-1 is located, is a frequently reported finding in leiomyosarcomas.32 t(10;17)(q22.1;p13) has been described as the sole genetic change that is observed in a uterine leiomyosarcoma.33 17p13 loss involving the p53 location was observed in two of the six cases of uterine leiomyosarcoma in this study, involving the associated leiomyoma-like area also in one case, and as an additional change in another case. Figure 3 summarizes the changes observed in leiomyoma-like areas and in uterine leiomyosarcoma. The findings of this study show that the gains of oncogenes and loss of tumor suppressor genes underlie the growth of uterine leiomyosarcomas, as has been shown previously.34
The presence of benign- and malignant-looking areas in a tumor could be explained in three different ways. The benign- and malignant-looking areas could be different tumors, in which case, they would be expected to show distinct genetic changes. The benign-looking areas could be better differentiated areas of the tumor, in which case, the genetic profile of these two different areas would be the same. Finally, malignant areas could arise from benign-looking areas. In this case, the two areas would share similar changes, but additional changes would be present in malignant areas. The findings of this study showed the last possibility as the underlying explanation for the presence of leiomyoma-like areas associated with uterine leiomyosarcoma areas examined in this study. In all six cases examined, genetic aberrations found in leiomyoma-like areas are also found in the corresponding uterine leiomyosarcoma areas. In addition, uterine leiomyosarcoma areas have additional genetic aberrations.
The large variation of the patterns of DNA changes from one uterine leiomyosarcoma to another suggests that the growth properties needed to develop the malignant phenotype in these tumors may result from a varied collection of DNA changes. Such varied changes are also indicative of genetic instability.
Although these DNA findings provide potential insight into the origins of uterine leiomyosarcoma, the readers should be aware that there may be additional, and potentially more important, changes that cause uterine leiomyosarcoma. Any balanced genetic alterations (such as a translocation), any mutations or any changes below the threshhold of detection of DNA CGH would not be picked up by this CGH study. Nevertheless, the persistence of DNA changes observed in the leiomyoma-like areas in the associated uterine leiomyosarcoma supports our hypothesis that uterine leiomyosarcoma may arise from associated leiomyoma-like areas.
The findings of this study should not be interpreted to mean that leiomyomas in general are precancerous lesions. As the frequency of leiomyosarcomas is only 0.1–0.3% of the frequency of leiomyomas,35 only rare leiomyomas progress to leiomyosarcoma. As cellular and symplastic leiomyoma-like areas were overrepresented in uterine leiomyosarcoma-associated leiomyoma-like areas, leiomyomas with this morphology may be more prone to malignant transformation than usual type leiomyomas.
The findings of this study may have implications for the origin of sarcomas in general. Although some sarcomas have been associated with specific genetic changes,36 very little is currently known about pre-neoplastic lesions for most sarcomas. Most malignant peripheral nerve sheath tumors are thought to arise by malignant transformation of neurofibromas,37 but DNA studies to confirm such progression have not been reported.
References
Jones MW, Norris HJ . Clinicopathologic study of 28 uterine leiomyosarcomas with metastasis. Int J Gynecol Pathol 1995;14:243–249.
Wilkinson N, Rollason TP . Recent advances in the pathology of smooth muscle tumours of the uterus. Histopathology 2001;39:331–341.
Major FJ, Blessing JA, Silverberg SG, et al. Prognostic factors in early-stage uterine sarcoma: a Gynecologic Oncology Group study. Cancer 1993;71:1702–1709.
Oliva E, Clement PB, Young RH . Mesenchymal tumours of the uterus: selected topics emphasizing diagnostic pitfalls. Curr Diagn Pathol 2002;8:268–282.
Hendrickson MR, Kempson RL . A diagnostic approach to smooth muscle tumours of the uterus. Curr Diagn Pathol 2000;6:21–30.
Clement PB, Young RH . Mesenchymal and mixed epithelial-mesenchymal tumors of the uterine corpus and cervix. In: Clement PB (eds) Atlas of Gynecologic Surgical Pathology. WB Saunders: Philadelphia, Pa, 2000, pp 177–210.
Longacre TA, Hendrickson MR, Kempson RL . Predicting clinical outcome for uterine smooth muscle neoplasms with a reasonable degree of certainty. Adv Anat Pathol 1997;4:95–104.
Mittal K, Popiolek D, Demopoulos RI . Uterine myxoid leiomyosarcoma within a leiomyoma. Hum Pathol 2000;31:398–400.
Mittal K, Joutovsky A . Areas with benign morphologic and immunohistochemical features are associated with some uterine leiomyosarcomas. Gynecol Oncol 2007;104:362–365.
Toruner GA, Streck DL, Dermody JJ, et al. An oligonucleotide based array-CGH system for detection of genome wide copy number changes including subtelomeric regions for genetic evaluation of mental retardation. Am J Med Genet 2007;143:824–829.
Jatoi N . Leiomyosarcoma: rare malignant change in a leiomyoma. J Coll Physicians Surg Pak 2003;13:106–107.
Kir G, Eren S, Akoz I, et al. Leiomyosarcoma of the broad ligament arising in a pre-existing pure neurilemmoma-like leiomyoma. Eur J Gynaecol Oncol 2003;24:505–506.
Scurry J, Hack M . Leiomyosarcoma arising in a lipoleiomyoma. Gynecol Oncol 1990;39:381–383.
Mittal K, Demopoulos RI . MIB-1 (Ki-67), p53, estrogen receptor, and progesterone receptor expression in uterine smooth muscle tumors. Hum Pathol 2001;32:984–987.
Bohmann D, Bos TJ, Admon A, et al. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 1988;238:1386–1392.
Wang XC, Tian J, Tian LL, et al. Role of Cks1 amplification and overexpression in breast cancer. Biochem Biophys Res Commun 2009;379:1107–1113.
Semba K, Nishizawa M, Miyajima N, et al. Yes-related protooncogene, syn, belongs to the protein-tyrosine kinase family. Proc Natl Acad Sci USA 1986;83:5459–5463.
Almoguera C, Shibata D, Forrester K, et al. Most human arcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988;53:549–554.
Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res 2006;12:1647–1653.
Sharrocks AD, Brown AL, Ling Y, et al. The ETS-domain transcription factor family. Int J Biochem Cell Biol 1997;29:1371–1387.
Ohta S, Uemura H, Matsui Y, et al. Alterations of p16 and p14ARF genes and their 9p21 locus in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2009;107:81–91.
Wong VC, Chan PL, Bernabeu C, et al. Identification of an invasion and tumor-suppressing gene, Endoglin (ENG), silenced by both epigenetic inactivation and allelic loss in esophageal squamous cell carcinoma. Int J Cancer 2008;123:2816–2823.
Kurup S, Abramsson A, Li JP, et al. Heparan sulphate requirement in platelet-derived growth factor B-mediated pericyte recruitment. Biochem Soc Trans 2006;34:454–455.
Zweemer RP, Ryan A, Snijders AM, et al. Comparative genomic hybridization of microdissected familial ovarian carcinoma: two deleted regions on chromosome 15q not previously identified in sporadic ovarian carcinoma. Lab Invest 2001;81:1363–1370.
Castells A, Gusella JF, Ramesh V, et al. A region of deletion on chromosome 22q13 is common to human breast and colorectal cancers. Cancer Res 2000;60:2836–2839.
Cho YL, Bae S, Koo MS, et al. Array comparative genomic hybridization analysis of uterine leiomyosarcoma. Gynecol Oncol 2005;99:545–551.
Dang CV, O'donnell KA, Juopperi T . The great MYC escape in tumorigenesis. Cancer Cell 2005;8:177–178.
Conti A, Aguennouz M, La Torre D, et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol 2009;93:325–332. [Epub ahead of print].
Borkosky SS, Gunduz M, Nagatsuka H, et al. Frequent deletion of ING2 locus at 4q35.1 associates with advanced tumor stage in head and neck squamous cell carcinoma. J Cancer Res Clin Oncol 2008;135:703–713. [Epub ahead of print].
Schmidt H, Semjonow A, Csiszar K, et al. Mapping of a deletion interval on 8p21-22 in prostate cancer by gene dosage PCR. Verh Dtsch Ges Pathol 2007;91:302–307.
Perner S, Wagner PL, Soltermann A, et al. TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J Pathol 2009;217:65–72.
Classon M, Harlow E . The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer 2003;2:910–917.
Dal Cin P, Boghosian L, Crickard K, et al. t(10;17) as the sole chromosome change in a uterine leiomyosarcoma. Cancer Genet Cytogenet 1988;32:263–266.
Packenham JP, du Manoir S, Schrock E, et al. Analysis of genetic alterations in uterine leiomyomas and leiomyosarcomas by comparative genomic hybridization. Mol Carcinog 1997;4:273–279.
Leibsohn S, d'Ablaing G, Mishell Jr DR, et al. Leiomyosarcoma in a series of hysterectomies performed for presumed uterine leiomyomas. Am J Obstet Gynecol 1990;62:968–974.
Osuna D, de Alava E . Molecular pathology of sarcomas. Rev Recent Clin Trials 2009;4:12–26.
Woodruff JM . Pathology of tumors of the peripheral nerve sheath in type 1 neurofibromatosis. J Med Genet (Semin Med Genet) 1999;89:23–30.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors have no financial interest in the contents of the manuscript.
Rights and permissions
About this article
Cite this article
Mittal, K., Chen, F., Wei, J. et al. Molecular and immunohistochemical evidence for the origin of uterine leiomyosarcomas from associated leiomyoma and symplastic leiomyoma-like areas. Mod Pathol 22, 1303–1311 (2009). https://doi.org/10.1038/modpathol.2009.96
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.96
Keywords
This article is cited by
-
Uterine fibroid-like tumors: spectrum of MR imaging findings and their differential diagnosis
Abdominal Radiology (2022)
-
Benign uterine mass—discrimination from leiomyosarcoma by a preoperative risk score: a multicenter cohort study
Archives of Gynecology and Obstetrics (2019)
-
Characterization of MED12, HMGA2, and FH alterations reveals molecular variability in uterine smooth muscle tumors
Molecular Cancer (2017)
-
Epigenetic and genetic landscape of uterine leiomyomas: a current view over a common gynecological disease
Archives of Gynecology and Obstetrics (2017)