Abstract
Hepatocellular carcinoma (HCC) represents the fifth and ninth cause of mortality among male and female cancer patients, respectively and typically arises on a background of a cirrhotic liver. HCC develops in a multi-step process, often encompassing chronic liver injury, steatosis and cirrhosis eventually leading to the malignant transformation of hepatocytes. Aberrant expression of the class I homeobox gene family (HOX), a group of genes crucial in embryogenesis, has been reported in a variety of malignancies including solid tumors. Among HOX genes, HOXA13 is most overexpressed in HCC and is known to be directly regulated by the long non-coding RNA HOTTIP. In this study, taking advantage of a tissue microarray containing 305 tissue specimens, we found that HOXA13 protein expression increased monotonically from normal liver to cirrhotic liver to HCC and that HOXA13-positive HCCs were preferentially poorly differentiated and had fewer E-cadherin-positive cells. In two independent cohorts, patients with HOXA13-positive HCC had worse overall survival than those with HOXA13-negative HCC. Using HOXA13 immunohistochemistry and HOTTIP RNA in situ hybridization on consecutive sections of 16 resected HCCs, we demonstrated that HOXA13 and HOTTIP were expressed in the same neoplastic hepatocyte populations. Stable overexpression of HOXA13 in liver cancer cell lines resulted in increased colony formation on soft agar and migration potential as well as reduced sensitivity to sorafenib in vitro. Our results provide compelling evidence of a role for HOXA13 in HCC development and highlight for the first time its ability to modulate response to sorafenib.
Similar content being viewed by others
Main
Hepatocellular carcinoma (HCC) represents the fifth and ninth cause of mortality among male and female cancer patients, respectively1, 2 and typically arises on a background of a cirrhotic liver.3 HCC develops in a multi-step process, often encompassing chronic liver injury, steatosis and cirrhosis eventually leading to the malignant transformation of hepatocytes.4 HCC is unique among cancers, in that no standard first-line cytotoxic therapy is routinely used in the clinic5 and limited therapeutic choices are available.6, 7 Currently, the multi-tyrosine kinase inhibitor sorafenib is the only option for HCCs in advanced stages. Of note, there are no validated predictive biomarkers for sorafenib response in HCC.8
Class I homeobox gene family (HOX) consists of a group of 39 highly conserved transcription factors known for their role as master regulators of embryonic development.9 The HOX genes are distributed over 4 chromosomal regions each containing between 9 and 11 members that aligned into 13 paralogous groups.10, 11 During developmental phases, HOX genes regulate the formation of distinct anatomical regions, through the maintenance of spatio-temporal collinearity across the anterior-posterior body axis.12, 13, 14 By contrast, in adult tissues, HOX genes maintain homeostasis by preserving the coordinates established during embryonic growth.15, 16
Aberrant expression of the HOX genes has been reported in a variety of malignancies including solid tumors.17, 18, 19, 20, 21 Among HOX genes, HOXA13 has been reported to be the most deregulated in HCC.22, 23, 24, 25 An earlier study found that knocking down HOXA13 reduces anchorage-dependent growth in vitro and cancer cell growth in mice with esophageal squamous cell carcinoma.26 Moreover, upregulation of HOXA13 has been linked to the activation of the Wnt and TGF-β pathways,21 suggesting that HOXA13 may have a role in determining the aggressiveness of HCCs. Furthermore, we recently reported that HOXA13 is directly regulated by the long non-coding RNA (lncRNA) HOXA transcript at the distal tip (HOTTIP), located in physical contiguity with HOXA13, via the interaction with the WDR5/MLL complex.22 Notably, HOXA13 and HOTTIP co-expression was found to be associated with disease progression, metastasis formation and worse clinical outcome,22 thus suggesting that the molecular axis controlled by HOXA13 and HOTTIP has a pivotal role in HCC progression.22
In this study, we sought to determine whether HOXA13 is expressed at the protein level and whether its expression is associated with prognosis. We also evaluated the effect of HOXA13 on sorafenib response in vitro using stable liver cancer cell lines with either overexpression or knockdown of HOXA13.
Materials and methods
Tissue Microarray Data and Patient Samples
Tissue microarray (TMA) was constructed from formalin-fixed and paraffin-embedded (FFPE) specimens using a custom-built instrument (Beecher Instruments, Silver Spring, MD, USA) as previously described.27 Specimens were mostly collected from patients with early stage disease who were eligible for and underwent surgical resection without prior treatment. The TMA contained 434 tissue specimens of which 305 were suitable for evaluation, which included 115 HCC, 82 cirrhotic liver, and 108 normal liver samples (Supplementary Table 1). Exclusion criteria from the analysis were the absence of tissue punch, poor HOXA13 immunostaining quality or the lack of comprehensive clinical data. Tissue samples were retrieved from the archives of the Institute of Pathology, University Hospital Basel, Switzerland. Histologic grading and classification of HCCs and non-neoplastic liver tissue samples was performed by two experienced pathologists with an expertise in liver pathology (MSM and LMT) according to the World Health Organization (WHO) guidelines28 and the Edmondson & Steiner grading system.29 Additionally, samples were classified using the Barcelona Clinic Liver Cancer (BCLC) staging classification.30 A second cohort of samples comprising whole sections from 43 resected HCC samples from sorafenib-treated patients was obtained from the Unit of Pathology, University of Milan and Humanitas Clinical and Research Center, Milan, Italy (Supplementary Table 2). Histologic grading and classification of this cohort of samples was performed by three experienced pathologists (CN, MR, and LMT) as described for the TMA cohort. The study was approved by the institutional review boards of the respective authors’ institutions.
Immunohistochemistry
Immunohistochemical (IHC) staining for HOXA13 and E-cadherin was performed on 4 μm sections of FFPE tissue using primary antibodies against HOXA13 (Abcam, Cambridge, UK; clone ab106503, dilution 1:200) and E-cadherin (Cell Marque, Rocklin, CA, USA; clone EP000Y, ready-to-use). Specifically, sections were pre-treated with CC1 (Ventana Medical Systems, Tucson, AZ, USA) and incubated with primary antibodies against HOXA13 and E-cadherin. Staining procedures were performed on a Benchmark immunohistochemistry staining system (Ventana Medical Systems) using iVIEW-DAB as chromogen as previously described.31 Positive and negative controls were included in each run. Immunoreactivity was scored by two experienced pathologists (MSM and LMT) For HOXA13, TMA specimens (tissue punches) were analyzed by evaluating the staining intensity (0–3, Figure 1a). For HOXA13 on whole tissue sections using at least two non-consecutive fields, both the staining intensity (0–3) and the percentage of stained cells (ie, hepatocytes) were evaluated as described by Allred et al.32 and classified as HOXA13 high and HOXA13 low using a semi-quantitative score accounting for both staining intensity and the percentage of stained cells using the following formula:
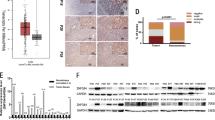
HOXA13 is expressed in a subset of aggressive HCCs and is inversely correlated with E-cadherin expression. (a) Representative micrographs of HOXA13 IHC with different staining intensities. (b) Percentages of HOXA13-positive and -negative normal, cirrhotic, and tumoral livers. (c) HOXA13 intensity in poorly, moderately and well differentiated HCCs. (d) E-cadherin expression levels in normal, cirrhotic, HOXA13-negative, and HOXA13-positive tumoral livers. (e) E-cadherin expression levels in normal, cirrhotic, and tumoral livers according to HOXA13 intensities. (f) E-cadherin and N-cadherin protein levels upon overexpression of HOXA13 in HepG2 cell lines. P-values≤0.05 were considered statistically significant. *P-values≤0.05; Error bars, s.d. of the mean.

where xi is the percentage of cells with intensity i. Sample with positive and negative scores were classified as HOXA13 high and HOXA13 low, respectively.
Analysis of Putative HOXA13 Binding Sites on CDH1 Promoter
The promoter sequence of the human CDH1 gene (encoding E-cadherin, Unigene ID: Hs.461086, ID of transcript: NM_004360) was retrieved from the DataBase of Transcriptional Start Site (DBTSS, http://dbtss-old.hgc.jp/hg19_mm9/), encompassing the genomic regions 2000 bp upstream to 400 bp downstream of the CDH1 transcriptional start site. The CDH1 promoter sequence was scanned for the presence of putative binding sites of HOXA13 using the JASPAR algorithm33 (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl) and the frequency matrix model (MA0650.1) corresponding to the human HOXA13 (Supplementary Figure 1a). The default relative score cut-off of 0.8 was used. Based on the results from JASPAR, a graphical representation of the distribution of putative binding sites of HOXA13 on the promoter region of CDH1 gene was obtained using ConSite service (http://consite.genereg.net/).
HOTTIP RNA In Situ Hybridization
Single color RNA in situ hybridization (ISH) was performed using the RNAscope® 2.0 HD Red Chromogenic Reagent Kit (Advanced Cell Diagnostics, Newark, CA, USA) per the manufacturer’s guidelines. Paired double-Z oligonucleotide probes were designed against Hs-HOTTIP RNA (Catalog number 400133, Advanced Cell Diagnostics) using custom software as described previously.34 GenBank accession numbers, number of probe pairs, and probe regions are: NR_037843, 20 pairs, nucleotides 388–1386. 4 μm sections of FFPE tissue were incubated at 60 °C for 1 h before use. After de-paraffinization and dehydration, the tissues were air-dried and treated with peroxidase blocker before boiling at 100–104 °C in a pre-treatment solution for 15 min. Protease was then applied at 40 °C for 30 min. Target probes were hybridized at 40 °C for 2 h. All hybridizations were performed in a HybEZ system (Advanced Cell Diagnostics) at 40 °C. Hybridization signals were detected by chromogenic precipitate development and RNA staining signal was identified as red dots. Following the RNAscope assay, samples were counterstained for 2 min with 50% Gill’s Hematoxylin diluted in dH2O. Each sample was quality controlled for RNA integrity with a probe specific to the housekeeping gene cyclophilin B (PPIB); only samples with an average of >4 dots per cell were included for analysis. Negative control background staining was evaluated using a probe specific to the bacterial dapB gene. To verify that the RNAscope method was performed with technical accuracy, references slides consisting of FFPE HeLa cells were tested with PPIB and dapB in parallel with tissue samples. Staining intensity was scored on the 4-point scale (ie, 0–3) by two experienced pathologists (MSM and LMT) as for HOXA13 IHC staining intensity.
Cell Lines Maintenance and Generation of Stable Clones
Liver cancer-derived cell lines SNU-449 and HepG2 were obtained from the Laboratory of Experimental Carcinogenesis (Bethesda, MD, USA), authenticated by short tandem repeat profiling as previously described35 and tested for mycoplasma infection using a PCR-based test (ATCC). All cell lines were maintained under the condition as recommended by the provider. Total RNA and proteins were extracted from cells at 75% confluence using TRIZOL (Invitrogen, Carlsbad, CA, USA) according to the manufacture’s guidelines. SNU-449 and HepG2 cells were transfected as described by Wang et al.36 to generate stable HOXA13-expressing cell lines.
Stable HOXA13 knockdown cells were obtained using the TALEN. Specifically, 13 × 104 cells were plated in 6-well plate and after 24 h cells were transfected with lipofectamine3000 (Thermo Fisher Scientific, Reinach, CH, USA) according to the manufacturer’s procedure with pcDNA3.1+ containing the full-length human cDNA of HOXA13 and empty vector as control.22 The cells were selected with 50 μg/ml of G418 (Gibco, Thermo Fisher Scientific, Reinach, CH, USA) for 72 h. For the knocking down of HOXA13, SNU-449, and HepG2 were simultaneously transfected with TAL2209/2209, a gift from Keith Joung (plasmids # 36672 and #36671, Addgene, Cambridge, MA, USA).37 24 h after transfection, cells were plated in 96-well plate using a dilution of 1 cell/well.
Relative Expression of HOXA13 by qRT-PCR
cDNA was synthesized from 1 μg of total RNA using Superscript II RNase H-reverse transcriptase (Invitrogen). HOXA13 expression was assessed using the TaqMan® Probe-Based Gene Expression Analysis (Applied Biosystems, Foster City, CA, USA; probe Hs00426284_m1). Measurements were normalized using GAPDH and 18S levels as reference. The fold change in gene expression was calculated using the standard ΔΔCt method as previously described.23, 38 All reverse transcriptase reactions, including no-template controls, were run on an Applied Biosystem 7900HT thermocycler. All samples were analyzed in triplicate.
Soft Agar Colony Assay, Proliferation Assay, and In Vitro Sorafenib Exposure
Soft agar colony formation assay was performed as previously described.39 Specifically, 103 cells were plated in 60 mm dishes in a solution containing Dulbecco’s modified Eagle’s medium 2X (Thermo Fisher Scientific, Reinach, CH, USA), TPB Buffer (Thermo Fisher Scientific, Reinach, CH, USA) and 1.25% of Noble Agar (Difco, Franklin Lakes, NJ, USA). Cells were harvested and counted, and a layer of 7 ml solution containing Noble Agar was left to polymerize on the bottom of the dishes. Meanwhile cells were diluted in 2 ml of the same solution, plated and left growing for 2 weeks in the incubator. Pictures of four different fields were taken using an inverted microscope and cell colonies were counted and measured in size using the ImageJ software (Version 1.49). Colonies were classified into three groups based on thresholds defined using the size distribution (arbitrary units) of the control group as small (first quartile), medium (2nd and 3rd quartiles) and large (4th quartile).
Cell proliferation was assayed using the xCELLigence system (RTCA, ACEA Biosciences, San Diego, CA, USA). Background impedance of the xCELLigence system was measured for 12 s using 50 μl of room temperature cell culture media in each well of E-plate 16. SNU-449 and HepG2 cells were grown and expanded in tissue culture flasks as previously described.40 After reaching 75% confluence, the cells were washed with PBS and detached from the flasks using a short treatment with trypsin/EDTA. 10 000 cells were dispensed into each well of an E-plate 16. Growth and proliferation of the cells were monitored every 15 min up to 72 h via the incorporated sensor electrode arrays of the xCELLigence system, using the RTCA-integrated software according to the manufacturer’s parameters.
In vitro sorafenib exposure experiments were carried out as described by Rothweiler et al.41 Briefly, cells were grown as above and 24 h after cell seeding, sorafenib at final concentration of 10 μM or 20 μM or DMSO (0.1% v/v) was added. Cell growth was assayed for up to 72 h. Data analysis and data normalization to the time before sorafenib/ DMSO addition was carried out using the RTCA Software 1.2.1.1002. Cell index values from three replicate experiments were normalized to DMSO, which was set as 100%.
Migration Assay
Migration assay was performed with CIM plates using the xCELLigence technologies (OLS OMNI Life Science, Basel, Switzerland). Stably transfected cells were detached and counted. 3 × 104 cells were plated in each well according to manufacturer’s instruction and migration was assessed 12, 24, and 36 h after seeding.
Statistical Analysis
For statistical analysis, the Chi-squared test (χ2-test) and Fisher's exact test for categorical variables and Mann–Whitney U/multiple t-tests/t-tests for continuous variables were used. Patient survival was assessed using the Kaplan–Meier method and analyzed using the log-rank test. Correlation tests were performed using Spearman correlation. All tests were two-sided and P-values≤0.05 were considered statistically significant. Analyses were performed using GraphPad 6.0 (Prism, La Jolla, CA, USA) and SPSS (version 24, IBM, San Francisco, CA, USA).
Results
HOXA13 Protein is Increased in Poorly Differentiated and More Aggressive HCCs
In a previous study, our group reported an upregulation of the RNA expression levels of the HOXA13 gene in the majority of HCCs analyzed.22 To corroborate and to extend this finding to the protein level, we investigated, by means of IHC on a TMA, HOXA13 levels on a large cohort of HCC (n=115), cirrhotic liver (n=108), and normal liver (n=82) specimens (Supplementary Table 1). Samples were evaluated according to their staining intensity (Figure 1a). Our analysis revealed that HOXA13 was expressed (ie, intensities 1, 2, or 3) in 10% (8/82) of normal liver, 18% (19/108) of cirrhotic liver, and 42% (48/115) of HCC specimens with a monotonic increase from normal liver tissues to cirrhotic tissues to HCCs (P<0.001, χ2-test, Figure 1b). Stratification based on the individual HOXA13 IHC intensity scores in HCCs showed that those with high HOXA13 expression (intensity 3) were associated with the highest proportion (29%) of poorly differentiated tumors (P<0.001, χ2-test, Figure 1c). Altogether, these results highlighted that: (i) HOXA13 positivity is more frequent in HCCs compared with normal and cirrhotic livers and (ii) high HOXA13 expression is associated with high tumor grade.
Considering our previous finding that HOXA13 expression was associated with metastasis in HCC,22 we evaluated the expression of E-cadherin, a master regulation of the metastatic process,42 in the same set of samples. We observed that HOXA13-positive HCCs had lower percentages of E-cadherin-positive cells compared with HOXA13-negative HCCs, cirrhotic liver and normal liver specimens (mean of positive cells 48% vs 65%, 68% and 69%, respectively, all P<0.05, Mann–Whitney U-tests, Figure 1d). Stratification of HCCs based on HOXA13 staining intensity scores showed that E-cadherin levels were inversely correlated with HOXA13 expression (all P<0.05, Mann–Whitney U-tests, Figure 1e).
To determine if there is a causal link between the inverse expression pattern of HOXA13 and E-cadherin, we performed in silico and in vitro experiments to determine if HOXA13 could directly regulate the expression of E-cadherin (encoded by CDH1). Using the JASPAR algorithm,33 we identified several putative HOXA13 binding sites on the CDH1 promoter sequence, in particular in the region 2000 to 500 bp upstream of the CDH1 transcriptional start site (Supplementary Figure 1), suggesting that HOXA13 may directly interact with the CDH1 promoter. Indeed, upon stable overexpression of HOXA13 in the HCC HepG2 cell line, we observed a decrease in E-cadherin and, conversely, an increase in N-cadherin expression (Figure 1f). In line with a previously published report,43 our in silico and in vitro experiments suggest a possible role for HOXA13 in the regulation of epithelial-to-mesenchymal transition in directly binding to E-cadherin.
Taken together, these results suggest that HOXA13 overexpression is associated with a more undifferentiated and aggressive tumor phenotype in HCC and we provided additional evidences that HOXA13 may regulate E-cadherin.
HOXA13 Protein Level is Associated with HCC Patient Survival
To explore whether HOXA13 protein levels were associated with the outcome of HCC patients as we previously demonstrated for HOXA13 mRNA levels,22 we examined the patient overall survival (OS) and disease-free survival (DFS) for our cohort of TMA HCC samples (n=115, Supplementary Table 1). Patients with HOXA13-positive HCC had shorter OS compared with those with HOXA13-negative disease (median 52 vs 65 months, P=0.03, log-rank test, Figure 2a). However, we found no statistical difference in DFS between patients with HOXA13-positive and HOXA13-negative HCC (median 16 vs 20 months, P=0.33, log-rank test, Supplementary Figure 2a). Similarly, comparing the OS and DFS stratified according to HOXA13 IHC staining intensities demonstrated that patients with HCCs with the highest IHC intensity (ie, 3) had the worst OS, but no difference was observed in DFS (P=0.04 and P=0.24, respectively, log-rank tests, Figure 2b and Supplementary Figure 2b). Focusing on patients with BCLC grade C HCC, as this is the only group eligible for sorafenib treatment, we found that patients with HOXA13-positive HCC had worse OS than those with HOXA13-negative disease (median 20 vs 24 months, P=0.05, log-rank test, Figure 2c).
High HOXA13 expression is associated with poor overall survival in patients with HCC. Overall survival of 115 HCC patients, (a) stratified into HOXA13 negative (staining intensity 0) and HOXA13 positive (staining intensity 1/2/3) or (b) stratified according to HOXA13 staining intensities. (c) Overall survival of 26 BCLC stage C HCC patients stratified into HOXA13 negative (staining intensity 0) and HOXA13 positive (staining intensity 1/2/3). (d) Overall survival of 43 HCC sorafenib-treated patients, stratified into HOXA13 negative (staining intensity 0) and HOXA13 positive (staining intensity 1/2/3). Statistical comparisons were performed using log-rank tests. P-values≤0.05 were considered statistically significant.
To extend these results to sorafenib-treated patients, we obtained whole tissue sections of an independent cohort composed of late-stage sorafenib-treated HCCs (Supplementary Table 2) and classified them as HOXA13 high and -low based on a semi-quantitative score, taking into account the staining intensity and the proportion of positive cells (Methods). In accordance with the results obtained from patients with untreated HCCs, sorafenib-treated patients with HOXA13-positive HCCs showed decreased OS compared with patients with HOXA13-negative disease (median 31 vs 41 months, P=0.05, log-rank test, Figure 2d).
These results suggest the HOXA13 protein expression in HCC is associated with OS in patients treated or untreated with sorafenib.
HOXA13 Expression Overlaps with HOTTIP in HCC Samples
Based on gene expression array data, we previously showed that HOXA13 overexpression in HCC is associated with concomitant HOTTIP upregulation.22 To determine whether HOXA13 is upregulated in the same population of cells as HOTTIP, we performed HOXA13 IHC and HOTTIP RNA in situ hybridization (ISH) on consecutive sections of 16 resected HCCs from the sorafenib-treated cohort (Supplementary Table 2). We found that both HOXA13 and HOTTIP were heterogeneously expressed in 71% of cases analyzed. Comparing the expression patterns, we observed that HOXA13 and HOTTIP were expressed in virtually the same populations of hepatocytes (Figure 3a) and that the percentages of HOXA13-positive and HOTTIP-positive cells demonstrated a significant correlation (r =0.91, P <0.0001, Spearman correlation test, Figures 3b and c).
HOXA13 and HOTTIP expression patterns in HCC. (a) Representative micrographs of HOXA13 IHC and HOTTIP ISH stained HCCs. (b) Heat map displaying the percentage of HOXA13-positive and HOTTIP-positive cells. (c) Spearman correlation analysis performed for the percentages of HOTTIP-positive and HOXA13-positive cells.
HOXA13 Overexpression Enhances Cell Aggressiveness In Vitro
In line with the observations that HOXA13 expression is increased in HCCs compared with non-tumoral livers and in poorly differentiated HCCs compared with moderately and well differentiated HCCs, our previous results demonstrated that transient in vitro HOXA13 overexpression in liver cancer cells resulted in increased proliferation whereas HOXA13 inhibition strongly reduced cell proliferation.22 Therefore, we sought to investigate if stable ectopic HOXA13 overexpression would have oncogenic properties in in vitro models. We generated liver cancer-derived cell line SNU-449 clones that stably overexpressed HOXA13, as verified by qRT-PCR (Supplementary Figure 3a). To test whether the cells that overexpressed HOXA13 displayed a more aggressive phenotype, we performed a colony formation assay on soft agar and evaluated the number and size of the resulting colonies (Figure 4a). Compared with the untransfected cell lines and the empty vector controls, SNU-449 overexpressing HOXA13 displayed larger colonies (P<0.05, Mann–Whitney U-test, Figure 4b) and a higher number of colonies per analyzed field (P<0.05, Mann–Whitney U-test, Figure 4c). Additionally, we performed a migration assay on SNU-449 and HepG2 stably overexpressing HOXA13 and observed that forced expression of HOXA13 resulted in increased cell migration (both P<0.05, Figures 4d and e) in both cell lines, thus supporting and corroborating our hypothesis that HOXA13 promotes epithelial-to-mesenchymal transition.
Effect of HOXA13 overexpression on cell transformation in in vitro models. (a) Representative micrographs of colonies, (b) distribution of sizes of the colonies, and (c) number of colonies per field in control SNU-449 cells and SNU-449 cells stably overexpressing HOXA13. (d,e) Migration potential in SNU-449 and HepG2 cell lines at 12, 24, and 36 h in stable HOXA13 overexpressing cells. All experiments were performed in triplicates. Error bars, s.d. of the mean. Statistical comparisons were performed using unpaired t-tests with Welch correction. P≤0.05 were considered statistically significant. *P ≤0.05; ns: not significant.
Taken together our results suggest that, in addition to increasing cell proliferation, HOXA13 overexpression also enhances cell aggressiveness and migration in vitro.
HOXA13 Level Modulates Sorafenib Sensitivity In Vitro
To provide a functional basis for our observation that sorafenib-treated patients with high HOXA13 levels in their HCCs had worse survival, we sought to investigate the impact of HOXA13 modulation in HCC-derived cell lines upon sorafenib exposure in vitro. In addition to the liver cancer-derived cell lines (SNU-449 and HepG2) with stable HOXA13 overexpression using pCMV-vector, we also generated two cell lines (SNU-449 and HepG2) with HOXA13 knockdown using TALEN, as verified by qRT-PCR (Supplementary Figure 3). To define the appropriate dose of sorafenib in vitro, we performed a titration experiment using untransfected SNU-449 and HepG2 (Supplementary Figure 4) and observed a significant inhibition of cell viability at 20 μM sorafenib compared with other concentrations (P<0.05, multiple t-test, Supplementary Figure 4). We then compared the proliferation indices of treated and untreated cells with and without HOXA13 overexpression using the xCelligence system up to 72 h after sorafenib exposure. In accordance with our previous results,22 in the absence of sorafenib, HOXA13-overexpressing cells displayed increased cell proliferation compared with the vector control (P<0.01, multiple t-test, Figures 5a/c). By contrast, sorafenib treatment significantly reduced cell proliferation in the vector control but had no effect in HOXA13-overexpressing cells (P<0.01 and P>0.05, respectively, multiple t-tests, Figures 5a/c).
HOXA13 modulates sorafenib response in HCC cells. Effect of sorafenib treatment in cells with (a, c) stable overexpression of HOXA13 (SNU-449 and HepG2) or (b,c) knockdown of HOXA13 (SNU-449 and HepG2) compared with the untransfected controls. Quantification was performed using a spectrophotometer. All experiments were performed in triplicates. Error bars, s.d. of the mean. Statistical comparisons were performed using Holm-Šídák-corrected multiple t-tests an. P≤0.05 were considered statistically significant. **P≤0.01.
Conversely, when we compared the untreated cells with and without HOXA13 knockdown, HOXA13-knockdown cells had lower cell proliferation than cells with TALEN control in both SNU-449 and HepG2 (both P<0.01, multiple t-tests, Figures 5b/d). On the other hand, upon sorafenib exposure, cell proliferation was significantly reduced in HOXA13 knockdown cells compared with the cells with TALEN control in both cell lines (both P<0.01, multiple t-tests, Figures 5b/d). Overall, these results provide evidence that overexpression of HOXA13 modulates sorafenib response in HCC in vitro models.
Discussion
In this study, we reported that HOXA13 protein level increased monotonically from normal liver to cirrhotic liver to HCC. We also observed that HOXA13-positive HCCs were preferentially poorly differentiated and had fewer E-cadherin-positive cells. In addition, we demonstrated that HOXA13 positivity is associated with worse overall survival, but not disease-free survival, which we confirmed in a second cohort consisting of sorafenib-treated patients. Furthermore, we showed that HOXA13 and HOTTIP were co-expressed in the same population of neoplastic hepatocytes in HCC. Finally, using liver cancer-derived cell lines, we demonstrated that HOXA13 expression enhances cell aggressiveness and modulates sorafenib response in vitro.
Based on gene expression data, we previously reported that HOXA13 is significantly upregulated in HCC.22, 23 In this work, by analyzing a large cohort of HCC samples (n=115), we extended our results to the protein level showing consistent HOXA13 overexpression in HCCs. This observation complements our previous results on the mRNA level,22, 23 is in line with the findings of another group on the protein level24 and further highlights that HOXA13 is deregulated in a subgroup of poorly differentiated (ie, high Edmondson grade) HCCs. Additionally, we observed that HOXA13-positive HCCs harbored fewer E-cadherin-positive cells. The loss of E-cadherin has been linked to the formation of metastases through the process of epithelial-to-mesenchymal transition44 and several groups recently reported that HOXA13 expression promotes cell invasive properties, for instance in glioma by activating the Wnt and TGF-β pathways.21 Our observation of HOXA13-positive HCCs displaying a more aggressive phenotype is thus in line with our previously described association between high HOXA13 level and metastatic incidence22 and the association with worse prognosis in terms of OS in these patients. HOXA13 positivity has been associated with unfavorable survival in HCCs24 as well as in other tumor entities such as esophageal squamous cell carcinoma,26, 45 gastric cancer,46 and pediatric acute lymphoblastic leukemia.18, 47, 48 In contrast to the previous study in which the authors found associations of HOXA13 expression with both OS and DFS,24 we found that HOXA13 expression was only associated with unfavorable OS, but not DFS, in two independent cohorts, one of which was a cohort of patients with sorafenib-treated, late-stage disease.
The results from our in vitro experiments provide functional evidence for the association observed between HOXA13 overexpression and the more aggressive and less differentiated phenotype in HCCs. We observed loss of E-cadherin upon HOXA13 overexpression and identified putative HOXA13 binding sites in the CDH1 promoter, suggesting that HOXA13 may have a direct role in the regulation of E-cadherin expression. Additionally, we observed increased colony formation and migration potential in cell lines overexpressing HOXA13, demonstrating that HOXA13 confers a more aggressive phenotype. In terms of sensitivity to sorafenib, forced expression of HOXA13 resulted in reduced sensitivity in vitro whereas knockdown of HOXA13 had the opposite effect. Given the role of HOXA13 in regulating E-cadherin and the previously published results49 suggesting that epithelial-to-mesenchymal transition has a pivotal role in modulating drug resistance, one can speculate that HOXA13 may affect sensitivity to sorafenib through the regulation of the epithelial-to-mesenchymal transition process. Indeed, the reduced sensitivity to sorafenib may be related to the loss of E-cadherin and hence an epithelial phenotype. Conversely, upregulation of HOXA13 may also affect drug response to multiple drugs in multiple cancer types as HOXA13 overexpression has also been reported to result in gemcitabine resistance in pancreatic cancer.43 These results provide a functional basis for the suboptimal response of the subset of patients with HOXA13-positive disease and the observed reduced OS in this group of patients.
Several studies previously demonstrated that HOXA13 and HOTTIP expression are highly correlated.20, 50, 51, 52 Specifically, we reported that HOTTIP activates HOXA13 through epigenetic mechanisms including DNA methylation and histone modifications22 and Wang et al. described that, in the Cs12 gastric cells, HOTTIP knockdown attenuates the recruitment of WDR5 and MLL1 and promotes DNMT3B localization on the HOXA13 promoter.20 Here by analyzing HOXA13 IHC and HOTTIP ISH on consecutive HCC sections, we showed that HOXA13 and HOTTIP displayed intra-tumor heterogeneous pattern of expression and their concomitant overexpression was found in the same populations of transformed hepatocytes. To the best of our knowledge, this is the first in situ characterization of the pattern of HOTTIP expression in human tissues. Given the increased recognition of the role of lncRNAs in cancer,53 this observation and the technique are of significant interest. We foresee that in situ characterization of HOTTIP expression will enable the identification of the specific subpopulations of transformed or malignant cells that express HOTTIP.
There are several limitations to our study. Our first and larger cohort of resected samples consisted of TMA punches rather than whole sections, and thus the observed expression may not be representative of the individual tissue samples. To account for intra-tumor heterogeneous expression of HOXA13, for the second cohort for which whole sections were available, we used a semi-quantitative score that accounts for both staining intensity and the percentage of cells for each staining intensity. In addition, the number of patients with BCLC stage C disease and late-stage sorafenib-treated patients for survival analysis was small. It should be noted that, despite the small cohorts, we saw a consistent association of HOXA13 overexpression with worse prognosis in both cohorts. It should also be noted that access to such collections of specimens with the associated outcome data is extremely limited. Finally, our results have not provided mechanistic insight into the association of HOXA13, HOTTIP and E-cadherin expression and the effect of HOXA13 expression on sorafenib sensitivity. Although we found that HOTTIP and HOXA13 are expressed in same population of cells, we have not provided direct evidence to support the hypothesis that HOTTIP regulates HOXA13 expression. Due to the cis-regulatory mechanism of HOTTIP, the physical proximity of HOTTIP to the HOX genes (including HOXA13) on the genome is essential for its regulatory activity. Indeed, it has been demonstrated by,36 that ectopic HOTTIP expression driven by retroviral insertion sites scattered randomly throughout the genome is not sufficient to induce 5′ HOXA genes. Future experiments using genome editing technology to investigate the regulatory role of HOTTIP would be of great interest.
To conclude, using a combination of clinico-pathologic and protein expression data obtained from two independent cohorts of HCC patients, as well as in vitro experiments, we provided evidence for a role of the molecular axis comprising HOXA13/HOTTIP in HCC development and response to sorafenib.
References
Thorgeirsson SS, Grisham JW . Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002;31:339–346.
El-Serag HB, Rudolph KL . Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557–2576.
Mair RD, Valenzuela A, Ha NB et al. Incidence of hepatocellular carcinoma among US patients with cirrhosis of viral or nonviral etiologies. Clin Gastroenterol Hepatol 2012;10:1412–1417.
Farazi PA, DePinho RA . Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674–687.
Villanueva A, Hernandez-Gea V, Llovet JM . Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol 2013;10:34–42.
Forner A, Llovet JM, Bruix J . Hepatocellular carcinoma. Lancet 2012;379:1245–1255.
Worns MA, Galle PR . HCC therapies—lessons learned. Nat Rev Gastroenterol Hepatol 2014;11:447–452.
Cho JY, Paik YH, Lim HY et al. Clinical parameters predictive of outcomes in sorafenib-treated patients with advanced hepatocellular carcinoma. Liver Int 2013;33:950–957.
Graham A, Papalopulu N, Krumlauf R . The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989;57:367–378.
Scott MP . Vertebrate homeobox gene nomenclature. Cell 1992;71:551–553.
Krumlauf R . Hox genes in vertebrate development. Cell 1994;78:191–201.
Gehring WJ, Hiromi Y . Homeotic genes and the homeobox. Annu Rev Genet 1986;20:147–173.
Shaut CA, Keene DR, Sorensen LK et al. HOXA13 Is essential for placental vascular patterning and labyrinth endothelial specification. PLoS Genet 2008;4:e1000073.
de Laat W, Duboule D . Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 2013;502:499–506.
Soshnikova N, Duboule D . Epigenetic temporal control of mouse Hox genes in vivo. Science 2009;324:1320–1323.
Mallo M, Alonso CR . The regulation of Hox gene expression during animal development. Development 2013;140:3951–3963.
Cillo C, Cantile M, Faiella A et al. Homeobox genes in normal and malignant cells. J Cell Physiol 2001;188:161–169.
Bhatlekar S, Fields JZ, Boman BM . HOX genes and their role in the development of human cancers. J Mol Med 2014;92:811–823.
Kelly Z, Moller-Levet C, McGrath S et al. The prognostic significance of specific HOX gene expression patterns in ovarian cancer. Int J Cancer 2016;139:1608–1617.
Wang SS, Wuputra K, Liu CJ et al. Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget 2016;7:36049–36064.
Duan R, Han L, Wang Q et al. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-beta pathways. Oncotarget 2015;6:27778–27793.
Quagliata L, Matter MS, Piscuoglio S et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014;59:911–923.
Cillo C, Schiavo G, Cantile M et al. The HOX gene network in hepatocellular carcinoma. Int J Cancer 2011;129:2577–2587.
Pan TT, Jia WD, Yao QY et al. Overexpression of HOXA13 as a potential marker for diagnosis and poor prognosis of hepatocellular carcinoma. Tohoku J Exp Med 2014;234:209–219.
Tsang FH, Au SL, Wei L et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int 2015;35:1597–1606.
Gu ZD, Shen LY, Wang H et al. HOXA13 promotes cancer cell growth and predicts poor survival of patients with esophageal squamous cell carcinoma. Cancer Res 2009;69:4969–4973.
Baumhoer D, Tornillo L, Stadlmann S et al. Glypican 3 expression in human nonneoplastic, preneoplastic, and neoplastic tissues: a tissue microarray analysis of 4,387 tissue samples. Am J Clin Pathol 2008;129:899–906.
Bosman FT, Carneiro F, Hruban RH et al. WHO Classification of Tumours of the Digestive System. World Health Organization, 2010.
Edmondson HA, Steiner PE . Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 1954;7:462–503.
Llovet JM, Bru C, Bruix J . Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 1999;19:329–338.
Piscuoglio S, Lehmann FS, Zlobec I et al. Effect of EpCAM, CD44, CD133 and CD166 expression on patient survival in tumours of the ampulla of Vater. J Clin Pathol 2012;65:140–145.
Allred DC, Harvey JM, Berardo M et al. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Modern Pathol 1998;11:155–168.
Mathelier A, Fornes O, Arenillas DJ et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2016;44:D110–D115.
Wang F, Flanagan J, Su N et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22–29.
Weigelt B, Warne PH, Downward J . PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene 2011;30:3222–3233.
Wang KC, Yang YW, Liu B et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120–124.
Reyon D, Tsai SQ, Khayter C et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012;30:460–465.
de Kok JB, Roelofs RW, Giesendorf BA et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 2005;85:154–159.
Quintavalle C, Garofalo M, Zanca C et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene 2012;31:858–868.
Andreozzi M, Quintavalle C, Benz D et al. HMGA1 expression in human hepatocellular carcinoma correlates with poor prognosis and promotes tumor growth and migration in in vitro models. Neoplasia 2016;18:724–731.
Rothweiler S, Dill MT, Terracciano L et al. Generation of a murine hepatic angiosarcoma cell line and reproducible mouse tumor model. Lab Invest 2015;95:351–362.
Wendt MK, Taylor MA, Schiemann BJ et al. Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol Biol Cell 2011;22:2423–2435.
Li Z, Zhao X, Zhou Y et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med 2015;13:84.
Marcucci F, Stassi G, De Maria R . Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov 2016;15:311–325.
Ma RL, Shen LY, Chen KN . Coexpression of ANXA2, SOD2 and HOXA13 predicts poor prognosis of esophageal squamous cell carcinoma. Oncol Rep 2014;31:2157–2164.
Han Y, Tu WW, Wen YG et al. Identification and validation that up-expression of HOXA13 is a novel independent prognostic marker of a worse outcome in gastric cancer based on immunohistochemistry. Med Oncol 2013;30:564.
Shiba N, Ichikawa H, Taki T et al. NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes Chromosomes Cancer 2013;52:683–693.
Starkova J, Zamostna B, Mejstrikova E et al. HOX gene expression in phenotypic and genotypic subgroups and low HOXA gene expression as an adverse prognostic factor in pediatric ALL. Pediatr Blood Cancer 2010;55:1072–1082.
Shibue T, Weinberg RA . EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–629.
Chang S, Liu J, Guo S et al. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol Rep 2016;35:3577–3585.
Ge Y, Yan X, Jin Y et al. MiRNA-192 [corrected] and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet 2015;11:e1005726.
Quagliata L, Andreozzi M, Kovac M et al. SH2D4A is frequently downregulated in hepatocellular carcinoma and cirrhotic nodules. Eur J Cancer 2014;50:731–738.
Huarte M . The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253–1261.
Acknowledgements
This study was funded by Swiss Cancer League (Oncosuisse) grants KLS-2867-08-2011 and KLS-3639-02-2015 to LMT; Swiss Cancer League (Oncosuisse) grant KFS-3995-08-2016 to S.P. Swiss Cancer League (Oncosuisse) grant KFS-3302-08-2013 to MSM; Associazione Italiana per la ricerca sul Cancro grant number 15437 to MR; Swiss National Science Foundation (Ambizione grant number PZ00P3_168165) to SP and (SNF 310030_159529) to LMT. CKYN is funded by the European Research Council Synergy grant 609883 (MERiC). Funding bodies had no role in the design of the study, collection, analysis and interpretation of the data or the writing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
In this study, the authors show that HOXA13 protein levels increase as normal liver progresses to cirrhotic liver to hepatocellular carcinoma (HCC). HOXA13-positive HCCs are preferentially poorly differentiated and have fewer E-cadherin-positive cells, and HOXA13 positivity is associated with worse overall survival. In vitro, HOXA13 expression enhances cell migration and modulates sorafenib responsiveness. HOXA13 may therefore be a potential predictive biomarker for sorafenib responsiveness.
Supplementary information
Rights and permissions
About this article
Cite this article
Quagliata, L., Quintavalle, C., Lanzafame, M. et al. High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Lab Invest 98, 95–105 (2018). https://doi.org/10.1038/labinvest.2017.107
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.107
This article is cited by
-
Curdione induces ferroptosis mediated by m6A methylation via METTL14 and YTHDF2 in colorectal cancer
Chinese Medicine (2023)
-
Hsa-microRNA-1249-3p/Homeobox A13 axis modulates the expression of β-catenin gene in human epithelial cells
Scientific Reports (2023)
-
HOXA13 promotes the proliferation, migration, and invasion of nasopharyngeal carcinoma HNE1 cells by upregulating the expression of Snail and MMP-2
Scientific Reports (2023)
-
Development and validation of a predictive model in diagnosis and prognosis of primary glioblastoma patients based on Homeobox A family
Discover Oncology (2023)
-
Long non-coding RNA HOTTIP exerts an oncogenic function by regulating HOXA13 in nasopharyngeal carcinoma
Molecular Biology Reports (2023)