Abstract
Epithelial cell rests of Malassez (ERM) are quiescent epithelial remnants of the Hertwig’s epithelial root sheath (HERS) that are involved in the formation of tooth roots. ERM cells are unique epithelial cells that remain in periodontal tissues throughout adult life. They have a functional role in the repair/regeneration of cement or enamel. Here, we isolated odontogenic epithelial cells from ERM in the periodontal ligament, and the cells were spontaneously immortalized. Immortalized odontogenic epithelial (iOdE) cells had the ability to form spheroids and expressed stem cell-related genes. Interestingly, iOdE cells underwent osteogenic differentiation, as demonstrated by the mineralization activity in vitro in mineralization-inducing media and formation of calcification foci in iOdE cells transplanted into immunocompromised mice. These findings suggest that a cell population with features similar to stem cells exists in ERM and that this cell population has a differentiation capacity for producing calcifications in a particular microenvironment. In summary, iOdE cells will provide a convenient cell source for tissue engineering and experimental models to investigate tooth growth, differentiation, and tumorigenesis.
Similar content being viewed by others
Main
Tooth development is a highly orchestrated process that begins with the defined placement of individual teeth of specific shapes and sizes within the jaw. Signaling molecules produced by epithelial and mesenchymal cells during tooth development allow them to interact.1 Teeth are composed of three different mineralized tissues: cementum, dentin, and enamel. Enamel, which is the hardest substance in the body, is the only epithelial-derived calcified tissue in vertebrates. Enamel is generated by ameloblasts. Enamel matrix proteins (EMPs) and proteinases are important for enamel development (amelogenesis). Amelogenin, enamelin, and ameloblastin (AMBN) are the three major structural proteins in the enamel matrix of developing teeth.2 Amelogenin and AMBN are the major EMPs responsible for mineralizing enamel. Recently, two novel EMPs, amelotin (AMTN) and odontogenic ameloblast-associated protein (ODAM) were identified by a secretome analysis of the epithelial cells responsible for creating tooth enamel.3, 4 ODAM and AMTN are observed in the junctional epithelium as well as during the maturation stage of amelogenesis, suggesting their involvement in the formation and regeneration of junctional epithelium.3, 4, 5, 6
Hertwig’s epithelial root sheath (HERS) is involved in the induction of odontoblast differentiation and subsequent dentin deposition during root formation via epithelial–mesenchymal interactions.7 Epithelial cell rests of Malassez (ERM) are derived from HERS fragments during root development and are located in the periodontal ligament (PDL) tissues.8, 9, 10, 11, 12 ERM cells remain in PDL tissues throughout adult life, where they maintain homeostasis of the periodontium through reciprocal interactions with other periodontal cells. Moreover, ERM cells have roles in regeneration and periodontal maintenance through the expression of extracellular matrix proteins and growth factors.13, 14 The capability of ERM to produce bone/cementum-related proteins, such as alkaline phosphatase (ALP), osteopontin and bone sialoprotein (BSP), suggests that ERM is associated with cementum development.13, 15 Interestingly, ERM cells subcultured with dental pulp cells can differentiate into ameloblast-like cells and generate enamel-like tissues during crown formation.16 Indeed, HERS and ERM cells can produce proteins secreted by ameloblasts, such as EMPs under certain conditions.16, 17, 18, 19, 20, 21, 22 In addition, ODAM, AMTN, AMBN and amelogenin are produced by HERS cells entrapped in cementum but not by HERS cells along the developing root.6 ODAM is not expressed in ERM from young rodents and is only weakly and sporadically expressed in ERM from older rodents.6, 23 It has recently been shown that primary cultured HERS/ERM cells contain a population of primitive stem cells that express epithelial and embryonic stem cell markers.24 Indeed, ERM cells have a differentiation capacity to form bone, fat, cartilage, and neural cells in vitro, and they form bone, cementum-like structures, and Sharpey’s fiber-like structures in vivo.25 These findings suggest that a stem cell population exists in HERS/ERM. However, the nature of stem cells in ERM is not fully understood.
Recently, the generating or regenerating teeth using various bioengineering methods and cell-based approaches has been suggested as the next-generation therapy in the field of dentistry. Various types of epithelial and mesenchymal cells are used for generating bioengineered teeth by reconstituting the epithelial–mesenchymal interaction. The successful creation of a bioengineered tooth is achievable only when the odontogenic epithelium is reconstructed to produce a replica of natural enamel. Human odontogenic epithelium is lost after enamel has formed. Therefore, although dental mesenchymal stem cells have been well studied, a few studies have investigated odontogenic epithelial stem cells. A recent review described that odontogenic epithelial cells obtained from active dental lamina during the postnatal period, which are remnants of dental lamina, ERM, and reduced enamel epithelium, may be untapped sources of odontogenic epithelial stem cells.26 Indeed, epithelial cells obtained from tooth germ cells are frequently used to generate bioengineered teeth.27, 28, 29, 30, 31, 32 Surprisingly, gingival epithelial cells can be used to form bioteeth with embryonic tooth-inducing mesenchymal cells.33 Moreover, ERM cells regenerate enamel-like tissue when co-cultured with mesenchymal cells derived from the pulp,16 indicating that a possibility of generating bioteeth. Thus, regenerative medicine is being increasingly studied in dentistry, and a possible clinical application of bioengineered teeth is expected. However, the nature of stem cells in ERM must be understood to apply odontogenic epithelial cells for regenerative medicine in dentistry. In this study, we isolated human odontogenic epithelial cells from ERM in periodontal tissue and obtained iOdE cells. We then examined the molecular biological features of these cells and their possible application in tooth regeneration.
MATERIALS AND METHODS
Primary Culture and Subculture of Odontogenic Epithelial Cells
This study was approved by the Ethic Committee of Hiroshima University, and was performed in compliance with the Declaration of Helsinki. The experiments were performed in accordance with the approved guidelines. Tissue materials were treated using anonymization methods in a linkable fashion. At the time of tissue sampling, we explained to the volunteers about the use of their tissues in future research and they complied. Informed consent was obtained verbally, and their signatures were obtained.
Periodontal tissue fragments were collected from the wisdom teeth of volunteers at Hiroshima University Hospital. To avoid contaminating the experimental material with gingival tissues, the soft tissues attached to the cervical area of the tooth were carefully removed after extraction. The tooth with PDL was rinsed once in phosphate-buffered saline (PBS) and then immersed in a digestive solution containing 2 mg/ml collagenase and 0.25% trypsin at 37 °C for 1 h. Thereafter, the solutions were centrifuged to collect the released PDL cells. A few days after plating, epithelial-like cells with a cobblestone appearance were observed among the PDL cells. Contaminated epithelial cells were isolated from cultured human PDL cells using a limiting dilution method and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, St Louis, MO, USA) with 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, USA) and 100 U/ml penicillin–streptomycin (Gibco BRL) under conditions of 5% CO2 in air at 37 °C. The epithelial cells bypassed senescence and grew for more than 100 population doublings (PDs). Thereafter, we cultured the odontogenic epithelial cells in the same medium and analyzed it.
Cell Culture and Cell Growth Assay
The HSC2 oral squamous cell carcinoma cell line and the SaOS-2 human osteosarcoma cell line were provided by the Japanese Cancer Research Resources Bank and were routinely maintained in RPMI-1640 (Sigma-Aldrich) supplemented with 10% FBS and 100 U/ml penicillin–streptomycin under conditions of 5% CO2 in air at 37 °C. A human immortalized PDL cell line (HPL) and a human cementoblast-like cell line (HCEM1) were maintained in α-MEM (Sigma-Aldrich) supplemented with 10% FBS and 100 U/ml penicillin–streptomycin under 5% CO2 in air at 37 °C.34 The AM-1-immortalized ameloblastoma cell line was provided by Dr Hidemitsu Harada (Iwate Medical University).35 The OBA-9 immortalized human gingival epithelial cell line and AM-1 cells were maintained in Keratinocyte-SFM (Invitrogen, San Diego, CA, USA) under 5% CO2 in air at 37 °C. The HaCaT immortalized human keratinocyte cell line was obtained from Dr NE Fusenig (affiliated with the German Cancer Research Center). Normal human dermal fibroblasts (NHDF) were obtained from Lonza (Basel, Switzerland). The mesenchymal odontoma (mOd) cell line was established previously by us.36 HaCaT, NHDF and mOd cells were maintained in DMEM supplemented with 10% FBS and 100 U/ml penicillin–streptomycin under 5% CO2 in air at 37 °C.
The cells were seeded in 24-well plates at a density of 3000 cells per well and pre-cultured in the culture medium in a humidified atmosphere of 5% CO2 at 37 °C for 24 h for the cell growth assay. After pre-culture, the medium was replaced with fresh medium, and the number of trypsinized cells was counted using a cell counter (Coulter Z1; Coulter, Hialeah, FL, USA) after 0, 2, 4, and 6 days.
Histological Analysis
Thickened dental follicle tissue samples were retrieved from the Surgical Pathology Registry of Tokushima University Hospital. Three cases of thickened dental follicles were used in this study. This study was approved by the Ethics Committee of Tokushima University. The experiments were performed in accordance with the approved guidelines. Informed consent was obtained verbally from all patients, and their signature was obtained. Tissue fragments were fixed in 3.7% neutral-buffered formaldehyde solution and then embedded in paraffin. Thereafter, 4.5-μm sections were prepared on silicon-coated glass slides and stained with hematoxylin–eosin for histological examination. Other sections were used for von Kossa staining as follows. After several rinses with distilled water, the sections were incubated with 1% silver nitrate solution in a clear glass Coplin jar and placed under ultraviolet light for 20 min. The sections were then rinsed several times with distilled water again, and the unreacted silver was removed using 5% sodium thiosulfate for 5 min. After rinsing again, the sections were counterstained with nuclear fast red for 5 min.
Immunohistochemistry
The cells were cultured in 60-mm2 culture dishes until sub-confluent, fixed in 3.5% neutral-buffered formalin for 1 h, and washed three times with PBS. Thereafter, they were treated with 0.3% hydrogen peroxide in 100% methanol for 20 min and washed three times with PBS. Immunostaining was performed using the Envision System (Dako, Carpentaria, CA, USA). Pan-cytokeratin monoclonal antibodies (AE1/AE3, Biomeda, Foster City, CA, USA; CAM5.2, BD Immunocytometry System, San Jose, CA, USA), p63 monoclonal antibody (Dako), vimentin monoclonal antibody (Dako), α-smooth muscle actin (αSMA) monoclonal antibody (Dako), sclerostin polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and STEM121 monoclonal antibody (Takara Bio, Shiga, Japan) were applied at suitable dilutions and incubated overnight at 4 °C. Diaminobenzidine tetrahydrochloride was used as a chromogen and Mayer’s hematoxylin was used as a counterstain.
Sphere Culture
iOdE cells were cultured in plastic culture plates with a non-adhesive surfaces (Corning, Corning, NY, USA). The cells were plated at a density of 1 × 104 cells per well (6-well plates), and the culture medium was changed every other day until spheres formed.
Microarray Analysis
Total RNA from iOdE and OBA-9 cells was isolated using an RNeasy Mini kit (Qiagen, Valencia, CA, USA). RNA quality was first checked for chemical purity using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and then assessed for RNA integrity using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Total RNA (100 ng) was amplified and labeled using the Affymetrix Whole-Transcript Sense Target Labeling Protocol, and the labeled RNA was hybridized to the GeneChip Human Gene 2.0 ST Array (Affymetrix, Santa Clara, CA, USA). Data visualization and analysis were performed using GeneSpring GX (Version 12.1) software. Functional grouping and pathway analysis was performed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA). The accession number for the microarray dataset reported in this study is GSE75085 (Supplementary Tables S1 and S2).
Reverse Transcription PCR and Quantitative RT-PCR Analysis
Total RNA was isolated from culture cells using the RNeasy Mini kits (Qiagen). The RNA quality was first checked for chemical purity using a NanoDrop spectrophotometer. cDNA was synthesized from 1 μg total RNA using a PrimeScript RT reagent kit (Takara Bio). The primers used for reverse transcription PCR (RT-PCR) analysis are listed in Supplementary Table S3. Aliquots of total cDNA were amplified with Go Taq Green Master Mix (Promega, Madison, WI, USA), and amplifications were performed using the T100 thermal cycler (Bio-Rad, Richmond, CA, USA) for 30 cycles after initial denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, and extension for 1 min at 72 °C for all primers. Transcript levels were determined using a 7300 Real-Time PCR System (Life Technologies, Carlsbad, CA, USA) with SYBR Premix Ex Taq (Takara Bio). The relative mRNA expression of each transcript was normalized against GAPDH mRNA. The t-test was used to compare results between two groups. A P-value of <0.05 was considered significant.
Total RNA (1 μg) was used to synthesis cDNA with a RT2 First Strand Kit (Qiagen) to comprehensively analyze the expression of stem cell-related genes. The Human Stem Cell RT2 PCR Array (PAHS-405Z; Qiagen) was used to analyze the expression of genes related to stem cells in iOdE and OBA-9 cells. A complete list of the genes analyzed is presented in Supplementary Table S4. The relative quantities of gene-specific mRNAs were calculated using the 2−(ΔΔCt) method and web-based software available at the SABiosciences (Qiagen) website.
Mineralization Assay
Mineral nodule formation was detected using Dahl’s method for calcium. iOdE cells with or without mOd cells were plated in a 6-well plate and cultured in αMEM containing 10% FBS, 50 μg/ml ascorbic acid, 10 mM glycerophosphate, and 10 mM dexamethasone at 37 °C for 4 weeks. NHDF cells were used as a negative control. The cells were stained with Alizarin Red-S (ALZ).
Xenograft Assays
To examine whether iOdE cells generated a calcified focus in vivo, 1 × 106 iOdE cells in 50 μl PBS were mixed with 50 μl Matrigel (1 mg/ml; BD Matrigel Basement Membrane Matrix). Then, 100 μl of a mixture containing 1 × 106 iOdE cells were injected subcutaneously into multiple sites in severe combined immunodeficiency (SCID) mice (CREA Japan, Tokyo, Japan). This study was conducted in accordance with the ‘Fundamental Guidelines for the Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions’ under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government. The study protocols were approved by the Committee on Animal Experiments of Tokushima University. The animals were monitored weekly for mass formation and were sacrificed 1 month later.
RESULTS
Isolation of Odontogenic Epithelial Cells from ERM in Periodontal Tissue
We collected human PDL cells in periodontal tissue fragments from wisdom teeth using enzymatic digestion as reported previously.37 After plating, epithelial-like cells with a cobblestone appearance had contaminated the PDL cells; therefore, we isolated these epithelial-like cells using a limiting dilution method (Figure 1). Some populations among these isolated cells spontaneously bypassed senescence and grew over 100 PDs. We obtained immortalized odontogenic epithelial cells and called them iOdE. iOdE cells had growth characteristics similar to those of the HaCaT keratinocyte and HSC2 oral squamous cell carcinoma cell lines (Supplementary Figure S1).
Isolation of odontogenic epithelial cells from epithelial cell rests of Malassez (ERM) in the periodontal ligament (PDL). Diagrammatic presentation of immortalized odontogenic epithelial cell isolation. Periodontal tissue fragments were obtained from wisdom teeth, and the soft tissues attached to the cervical area were carefully removed after extraction. The extracted tooth was immersed in a digestive solution at 37 °C for 1 h, and the solution was centrifuged to collect the human PDL cells released. A few days later, epithelial-like cells with a cobblestone appearance were observed in the PDL culture. The epithelial-like cells were isolated using a limiting dilution method. The figure shows periodontal tissue, cultured PDL cells, and isolated odontogenic epithelial cells. Arrows show ERM in periodontal tissue (D: dentin, AB: alveolar bone). Scale bars are shown.
We performed an immunohistochemical analysis to examine the characteristics of iOdE cells. The iOdE and HSC2 cells were positive for epithelial markers, such as AE1/AE3, CAM5.2, and p63, but negative for the mesenchymal markerαSMA (Figure 2a). Some cells within the iOdE and HSC2 cells were vimentin positive (Figure 2a). Because all cells within iOdE cells were confirmed to be keratin positive (AE1/AE3 and CAM5.2), the mesenchymal cells had not contaminated in iOdE cells. The immortalized PDL cell line (HPL) was vimentin and αSMA positive but not positive for AE1/AE3, CAM5.2, or p63. Thus, iOdE cells exhibited properties similar to those of epithelial cells. Thereafter, we examined the mRNA expression of EMPs, including amelogenin, AMBN, ODAM, and AMTN, by qRT-PCR to demonstrate the odontogenic features of iOdE cells. Previous reports have shown that HERS/ERM cells can produce EMPs under certain conditions.16, 17, 18, 19, 20, 21, 22 In addition, ODAM, AMTN, AMBN, and amelogenin are produced by HERS cells entrapped in cementum.6 We used a gingival epithelial cell line (OBA-9) and an ameloblastoma cell line (AM-1) as negative and positive controls for EMP expression, respectively. As expected, AM-1 cells expressed all EMPs, whereas OBA-9 cells did not (Figure 2b). The expression of AMBN, ODAM, and AMTN but not that of amelogenin was observed in iOdE cells. In particular, the ODAM expression level was much higher in iOdE cells than that in AM-1 cells (Figure 2b).
Characterization of immortalized odontogenic epithelial (iOdE) cells. (a) Immunohistochemical staining of epithelial markers (AE1/3, CAM5.2, and p63) and mesenchymal markers (vimentin and αSMA) in iOdE, HSC2, and the HPL human immortalized PDL cell line. Scale bar, 50 μm. (b) Expression of enamel matrix proteins (amelobalstin, amelobalstin (AMBN), odontogenic ameloblast-associated protein (ODAM), and amelotin (AMTN)) was examined in iOdE, OBA-9 human immortalized gingival epithelial cells, and AM-1 human immortalized ameloblastoma cells by real-time reverse transcription PCR. Results are presented as mean±s.d. *P<0.05.
Comparing the Gene Expression Profiles between iOdE and Gingival Epithelial Cells
To examine the characteristics of iOdE cells, we compared the gene expression profiles of iOdE with OBA-9 cells by microarray analysis. A total of 1344 genes were more than two-fold upregulated and 1448 genes were more than two-fold downregulated in iOdE cells compared with those in OBA-9 cells (Figure 3a). Moreover, 71 genes were more than 10-fold upregulated (Supplementary Table S1) and 54 genes were more than seven-fold downregulated in iOdE cells (Supplementary Table S2). AMTN, cadherin 11 (CDH11), contactin 1 (CNTN1), and protein tyrosine phosphatase receptor-type Z polypeptide 1 (PTPRZ1) were included among the 10-fold upregulated genes. CDH11 is expressed in osteoblasts and regulates stem cell fate.38 CNTN1 is expressed at the surface of oligodendrocyte precursor cells and is involved in their proliferation and differentiation.39 PTPRZ1 is a receptor of a variety of cell adhesion and matrix molecules including CNTN1, expressed during neurogenesis.40, 41 We confirmed the upregulation of these genes in iOdE cells by qRT-PCR (Figures 2b and 3b). AM-1 cells also expressed these genes at higher levels. In addition, IPA was used to identify the biological pathways, networks, and functions significantly altered in iOdE cells. The genes identified by microarray analysis were classified using the IPA software into a variety of biological processes, including cellular proliferation, cellular development, differentiation, and cell morphology. The two most significant molecular and cellular processes affected in iOdE cells were the transforming growth factor (TGF)-β and WNT signaling pathways (Supplementary Figure S2a and b).
Comparison of gene expression profiles between immortalized odontogenic epithelial (iOdE) cells and gingival epithelial cells. (a) The gene expression profiles of iOdE and OBA-9 cells were compared by microarray analysis. Scatter plot of OBA-9 cells (x axis) and iOdE cells (y axis). Green lines indicate two-fold upregulation and downregulation. (b) CDH11, CNTN, and PTPRZ1 expression was examined by real-time reverse transcription PCR in iOdE, OBA-9, and AM-1 cells. Results are presented as mean±s.d. *P<0.05.
Stem Cell-Like Features of iOdE Cells
iOdE cells generated spheroids in the low-binding plates (Figure 4a). Moreover, we examined the expression of human telomerase reverse transcriptase (hTERT), which is a catalytic subunit of the enzyme telomerase, in primary, 19 PDs, and >100 PDs of iOdE cells. Interestingly, primary, 19 PDs, and >100 PDs of iOdE cells showed high hTERT expression levels, indicating that primary iOdE cells may have a high telomerase activity. Then, we examined the expression profiles of 84 stem cell-related genes in iOdE and OBA-9 cells by qRT-PCR arrays (Supplementary Figure S3). Among them, we identified a difference of at least two-fold in the expression of 12 genes, including ALP1, bone morphogenetic protein 2 (BMP2), cyclin D1 (CCND1), CD44, cell division cycle 42 (CDC42), growth differentiation factor-3 (GDF3), membrane metallo-endopeptidase (MME), V-myc myelocytomatosis viral oncogene homolog (MYC), sex determining region Y-box 1 (SOX1), SOX2, TERT, and tubulin, beta 3 (TUBB3) in iOdE cells (Figure 4b).
Stem cell-like features in immortalized odontogenic epithelial (iOdE) cells. (a) Spheroid formation by iOdE cells in the low-binding plates. The spheroids were photographed after 2 weeks. Representative photographs of spheroids are shown (PC and hematoxylin–eosin staining). PC: phase contrast. Scale bar is shown. The figure is representative of at least three independent experiments (n=3). (b) Human telomerase reverse transcriptase (hTERT) expression was examined by real-time reverse transcription (RT)-PCR in HSC2, HaCaT, primary of iOdE, 19 PDs of iOdE, and >100 PDs of iOdE cells. (c) Relative expression levels of 84 stem cell-related genes between iOdE cells and OBA-9 cells were examined by qRT-PCR array. Among the 84 stem cell-related genes, the relative expression of at least two-fold upregulated genes including, ALP1, BMP2, CCND1, CD44, CDC42, GDF3, MME, MYC, SOX1, SOX2, TERT, and TUBB3, are shown.
Osteogenic Differentiation of iOdE Cells
To understand the multipotency of iOdE cells, we examined the osteogenic differentiation of iOdE cells using ALZ staining after culture in mineralization-inducing media. Interestingly, iOdE cells showed intense ALZ staining in vitro (Figure 5a and b). The mineralization activity of iOdE cells was detected with mOd cells, which is a human mesenchymal cell line derived from odontoma tissue. mOd cells are thought to be mesenchymal cells derived from dental papilla.36 Interestingly, iOdE cells co-cultured with mOd cells showed intense ALZ staining (Figure 5a and b). Although the expression levels of mineralization-related genes in iOdE cells were lower than those in cementoblastic, PDL, and osteosarcoma cells, the expression of mineralization-related genes, such as runt-related transcription factor 2, Msh homeobox 2, ALP, BSP, type I collagen, and osteocalcin, was observed in iOdE cells (Figure 5c). A previous study showed that ERM cells are capable of undergoing the epithelial–mesenchymal transition (EMT) under osteogenic conditions.25 Therefore, we examined the expression of EMT markers, such as E-cadherin, N-cadherin, vimentin, SNAI1, SNAI2, ZEB1, and ZEB2, in iOdE cells after culture in mineralization-inducing media. Under osteogenic conditions, iOdE cells were spindle shaped (Figure 5d), and N-cadherin and SNAI2 expression was upregulated after culture in mineralization-inducing media (Figure 5e). Downregulation of E-cadherin and upregulation of SNAI1 and ZEB2 were not observed, and vimentin and ZEB1 expression was not detected (data not shown). These findings suggest that EMT is partially induced in iOdE cells under osteogenic conditions.
Osteoblastic differentiation of immortalized odontogenic epithelial (iOdE) cells in vitro. (a) Alizarin Red-S (ALZ) staining in iOdE cells with or without mesenchymal odontoma (mOd) cells. NHDF was used as the negative control for the mineralization activity. Cells were cultured in mineralization-inducing media (medium supplemented with ascorbic acid, sodium β-glycerophosphate, and dexamethasone) for 4 weeks. The cells were then stained with ALZ. Each panel represents at least three independent experiments for each group (n=3). (b) Phase contrast and ALZ staining images of iOdE cells with or without mOd cells are shown. PC: phase contrast. Scale bars, 50 and 100 μm. (c) Expression of mineralization-related genes in iOdE cells. Expression of mineralization-related genes including runt-related transcription factor 2 (RUNX2), Msh homeobox 2 (Msx2), ALP, BSP, type I collagen (COLI), and osteocalcin (OCN) was examined by RT-PCR in iOdE, HCEM1 (human immortalized cementoblast-like cell line), HPL (human immortalized PDL cell line), and SaOS (human osteosarcoma cell line) cells. HCEM1, HPL, and SaOS are known to have mineralization ability. GAPDH was used as a loading control. (d) Phase contrast images of iOdE cells after 0 and 3 days of culture in mineralization-inducing media are shown. Scale bar, 100 μm. (e) Expression of EMT markers, such as E-cadherin, N-cadherin, SNAI1, SNAI2 and ZEB1 was examined by real-time RT-PCR in iOdE cells after 0, 1, and 3 days of culture with mineralization-inducing media.
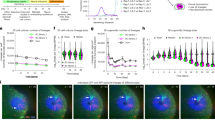
iOdE cells were injected with Matrigel subcutaneously into SCID mice to determine their mineralization activity in vivo. Interestingly, calcification foci were demonstrated by von Kossa staining in the transplanted iOdE cells (Figure 6a). We confirmed that the transplants in mice were derived from iOdE cells by staining with Stem121, which reacts specifically with a cytoplasmic protein in human cells (Figure 6a). Interestingly, we observed the expression of sclerosin as an osteocyte marker in some iOdE cells within transplants, indicating that some of these cells may differentiate into osteocytes (Figure 6a). Calcification foci are frequently found in surgically resected human thickened dental follicle tissues(Figure 6b; Supplementary Figure S4). Although calcification foci are not usually found in ERM, they are often seen in odontogenic epithelial islands within dental follicles (Figure 6b; Supplementary Figure S4). Although it remains unclear whether calcification foci were formed by odontogenic epithelial cells, we hypothesized that odontogenic epithelial cells may have the capacity for osteogenic differentiation in a certain microenvironment.
Osteoblastic differentiation of immortalized odontogenic epithelial (iOdE) cells in vivo. (a) Histology of iOdE cells in SCID mice. iOdE cells were injected subcutaneously into SCID mice. After 1 month, the mass was resected and histological analysis was performed. Calcification foci of iOdE cells are shown. Calcification foci were stained with von Kossa. Immunohistochemical staining for the human cytoplasmic marker, STEM121, and the osteocyte marker, sclerosin, in iOdE cells within transplants. Scale bar is shown. The figure is representative of at least three independent experiments (n=3). (b) Histology of odontogenic epithelial cells in human thickened dental follicle tissue. Calcification foci in odontogenic epithelial islands are shown. Calcification foci were stained with von Kossa. Scale bars are shown.
DISCUSSION
Here we obtained iOdE cells from contaminated epithelial cells within ERM in primary cultured human PDL cells. We confirmed that iOdE cells have odontogenic features, including the expression of EMPs. Because iOdE cells were spontaneously immortalized via by-passing senescence without any treatment, iOdE cells are thought to possess stem cell-like features. Indeed, iOdE cells showed an abnormal karyotype at late passage in culture (data not shown). It is well known that human embryonic stem cells lines and human induced pluripotent stem cells lines show an abnormal karyotype at late passage in culture.42 Moreover, human embryonic stem cells share cellular and molecular phenotypes with tumor cells and cancer cell lines, such as rapid proliferation rate, lack of contact inhibition, a propensity for genomic instability, high activity of telomerase, high expression of oncogenes such as MYC and KLF4, overall gene expression patterns, microRNA signatures, and epigenetic status.43 Therefore, an abnormal karyotype found in iOdE cells may indicate the stem cell-like properties.
In this study, iOdE cells formed spheroids and expressed stem cell-related genes. Moreover, iOdE cells showed mineralization activity in vitro and in vivo, and this mineralization activity was consistent with the following evidences; (i) calcification foci were often seen in odontogenic epithelial islands within dental follicles (Figure 6b; Supplementary Figure S4), and (ii) calcified areas resembling woven bone, osteodentin, and cementum were frequently observed in hyperplastic dental follicles.44 Moreover, Xiong et al.25 reported that subcutaneous transplantation of integrin α6/CD49f-positive ERM cells with hydroxyapatite tricalcium phosphate particles generates mineralized bone structures with osteoblast-like cells and osteocytes. In this study, calcification foci, but not mineralized bone structures, were observed in iOdE cells transplanted with Matrigel in vivo. However, differentiation into osteoblasts and the generation of mineralized bone structures may require bone substitutes, such as hydroxyapatite tricalcium phosphate particles. Thus, we established iOdE cells with stem cell-like features, suggesting that multipotent cells exist within ERM. Our observations are supported by the following previous findings; (i) HERS/ERM cells may differentiate into cementoblasts via the EMT;45, 46, 47, 48, 49 (ii) subcultured ERM cells combined with primary dental pulp cells seeded onto scaffolds become enamel-like tissues;16 and (iii) ERM demonstrates stem cell-like properties based on the differentiation capacity to form bone, fat, cartilage, and neural cells in vitro.25
In this study, we found that iOdE cells differentiated after treatment with osteogenic induction medium. As described above, ERM cells have a multipotency to form enamel, bone, fat, cartilage, and neural cells in vitro under certain conditions. Similar to tooth development, subcultured ERM cells combined with primary dental pulp cells seeded onto scaffolds revealed enamel-like tissues.16 Although the mechanism of cementoblast differentiation remains controversial, HERS/ERM cells may differentiate into cementoblasts via EMT.45, 46, 47, 48, 49 Moreover, ERM cells express EMT-related genes during osteogenic induction,25 and EMT is induced in an ERM cell line treated with TGF-β.49 Thus, we hypothesized that only some populations within HERS/ERM may have the capacity to differentiate into mesenchymal cells such as osteoblasts or cementoblasts, via EMT under particular conditions. Although EMT was not induced in iOdE cells using only TGF-β (data not shown), the IPA analysis revealed that the TGF-β and WNT signaling pathways were affected in iOdE cells (Supplementary Figure S1). Interestingly, iOdE cells became spindle shaped under osteogenic conditions (Figure 5d), and N-cadherin and SNAI2 expression was upregulated after culture in mineralization-inducing media (Figure 5e), suggesting that EMT is partially induced in iOdE cells under osteogenic conditions. In addition, we found that several growth factors affected the expression of mineralization-related genes in iOdE cells (Supplementary Figure S5). These findings suggest that growth factors from surrounding tissue and/or themselves are involved in the differentiation and function of stem cells within ERM. This observation is supported by the finding that signaling pathways mediated by several growth factors are involved in tooth morphogenesis.1 Moreover, stem cell populations within ERM may be involved in the pathogenesis of odontogenic cysts and tumors. Indeed, it is well known that ERM cells are associated with the pathogenesis of many odontogenic cysts and odontogenic tumors.50, 51
Odontogenic epithelial stem cells are involved in tooth development. They are of outer ectodermal origin and interact reciprocally with odontogenic mesenchymal stem cells. Dental lamina is the main source for odontogenic epithelial stem cells during tooth development. A recent review described that odontogenic epithelial stem cells obtained from active dental lamina during the postnatal period, remnants of dental lamina, remnants of dental lamina in the gubernaculum cord present above any erupting tooth, ERM, and reduced enamel epithelium may be untapped sources of odontogenic epithelial stem cells.26 ERM cells can be easily obtained from wisdom teeth or healthy premolar teeth using sequential enzymatic digestion.37 Therefore, ERM cells could be a useful stem cell source for regenerative medicine. However, a method to obtain a cell population with stem-like features from ERM cannot be established yet. In this study, we obtained spontaneously immortalized ERM cells with stem cell-like features after long-term culture, indicating that a cell population with features similar to stem cells exists in ERM. In this study, we obtained only one strain of iOdE cells. Indeed, immortalized odontogenic epithelial cells were obtained from ERM in pig (personal communication, Dr Yoshihiro Abiko). As we identified several highly expressed genes, such as ALP1, BMP2, CCND1, CD44, CDC42, GDF3, MME, MYC, SOX1, SOX2, TERT, TUBB3, AMTN, ODAM, CDH11, CNTN1, and PTPRZ1, in iOdE cells, these genes, particularly cell surface molecules, can be used to isolate stem cells within ERM. Interestingly, Xiong et al. reported that integrin α6/CD49f-positive ERM cells are multipotent. Further studies will be required to establish a method to isolate stem cells within ERM using specific molecules, including integrin α6/CD49f. To establish the method for isolating the stem cells within ERM requires further experiments. Generation or regeneration of teeth using various bioengineering methods and cell-based approaches has been suggested as next-generation therapy in the field of dentistry. Various types of epithelial and mesenchymal cells are used for generating bioengineered teeth by reconstituting the epithelial–mesenchymal interaction. Therefore, ERM with stem cell-like features can be used as odontogenic epithelial cells for generating bioengineered teeth. The nature of stem cells within ERM must be fully understood to apply ERM cells for regenerative medicine in dentistry. Therefore, iOdE will be a useful cell culture model to investigate tooth growth, differentiation, and tumorigenesis as well as understand the nature of ERM with stem cell-like features.
Accession codes
References
Thesleff I . Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci 2003;116:1647–1648.
Fincham AG, Moradian-Oldak J, Simmer JP . The structural biology of the developing dental enamel matrix. J Struct Biol 1999;126:270–299.
Moffatt P, Smith CE, St-Arnaud R et al. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochem J 2006;399:37–46.
Moffatt P, Smith CE, St-Arnaud R et al. Characterization of Apin, a secreted protein highly expressed in tooth-associated epithelia. J Cell Biochem 2008;103:941–956.
Nishio C, Warzen R, Moffatt P et al. Expression of odontogenic ameloblast-associated and amelotin proteins in the junctional epithelium. Periodontol 2000 2013;63:59–66.
Iwasaki K, Bajenova E, Somogyi-Ganss E et al. Amelotin—a novel secreted, ameloblast-specific protein. J Dent Res 2005;84:1127–1132.
Thomas HF, Kollar EJ . Differentiation of odontoblasts in grafted recombinants of murine epithelial root sheath and dental mesenchyme. Arch Oral Biol 1989;34:27–35.
Zeichner-David M, Oishi K, Su Z et al. Role of Hertwig's epithelial root sheath cells in tooth root development. Dev Dyn 2003;228:651–663.
Huang X, Bringas P Jr, Slavkin HC et al. Fate of HERS during tooth root development. Dev Biol 2009;334:22–30.
Hamamoto Y, Nakajima T, Ozawa H . Ultrastructural and histochemical study on the morphogenesis of epithelial rests of Malassez. Arch Histol Cytol 1989;52:61–70.
Loe H, Waerhaug J . Experimental replantation of teeth in dogs and monkeys. Arch Oral Biol 1961;3:176–184.
Sicher H . Periodontal ligament. In: Sicher H (ed). Orban’s Oral Histology and Embryology, 6th edn. The CV Mosby Company: St Louis, IL, USA, 1966, pp 176–196.
Bosshardt DD . Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J Dent Res 2005;84:390–406.
Thesleff I . Epithelial cell rests of Malassez bind epidermal growth factor intensely. J Periodontal Res 1987;22:419–421.
Rincon JC, Young WG, Bartold PM . The epithelial cell rests of Malassez—a role in periodontal regeneration? J Periodontal Res 2006;41:245–252.
Shinmura Y, Tsuchiya S, Hata K et al. Quiescent epithelial cell rests of Malassez can differentiate into ameloblast-like cells. J Cell Physiol 2008;217:728–738.
Bosshardt DD, Nanci A . Hertwig’s epithelial root sheath, enamel matrix proteins, and initiation of cementogenesis in procine teeth. J Clin Periodontol 2004;31:184–192.
Fong CD, Hammarstrom L . Expression of amelin and amelogenin in epithelial root sheath remnants of fully formed rat molars. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000;90:218–223.
Hamamoto CD, Nakajima T, Ozawa H et al. Production of amelogenin by enamel epithelium of Hertwig’s root sheath. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1996;81:703–709.
Luo W, Slavkin HC, Snead ML . Cells from Hertwig’s epithelial root sheath do not transcribe amelogenin. J Periodontal Res 1991;26:42–47.
Hasegawa N, Kawaguchi H, Ogawa T et al. Immunohistochemical characteristics of epithelial cell rests of Malassez during cementum repair. J Periodontal Res 2003;38:51–56.
Shimonishi M, Hatakeyama J, Sasano Y et al. In vitro differentiation of epithelial cells cultured from human periodontal ligament. J Periodontal Res 2007;42:456–465.
Nishio C, Wazen R, Kuroda S et al. Expression pattern of odontogenic ameloblast-associated and amelotin during formation and regeneration of the junctional epithelium. Eur Cell Mater 2010;20:393–402.
Nam H, Kim J, Park J et al. Expression profile of the stem cell markers in human Hertwig's epithelial root sheath/Epithelial rests of Malassez cells. Mol Cells 2011;31:355–360.
Xiong J, Mrozik K, Gronthos S et al. Epithelial cell rests of Malassez contain unique stem cell populations capable of undergoing epithelial-mesenchymal transition. Stem Cells Dev 2012;21:2012–2025.
Padma Priya S, Higuchi A, Abu Fanas S et al. Odontogenic epithelial stem cells: hidden sources. Lab Invest 2015;95:1344–1352.
Nakao K, Morita R, Saji Y et al. The development of a bioengineered organ germ method. Nat Methods 2007;4:227–230.
Ikeda E, Morita R, Nakao K et al. Fully functional bioengineered tooth replacement as an organ replacement therapy. Proc Natl Acad Sci USA 2009;106:13475–13480.
Yamamoto H, Kim EJ, Cho SW et al. Analysis of tooth formation by reaggregated dental mesenchyme from mouse embryo. J Electron Microsc (Tokyo) 2003;52:559–566.
Ohazama A, Modino SA, Miletich I et al. Stem-cell-based tissue engineering of murine teeth. J Dent Res 2004;83:518–522.
Duailibi MT, Duailibi SE, Young CS et al. Bioengineered teeth from cultured rat tooth bud cells. J Dent Res 2004;83:523–528.
Oshima M, Mizuno M, Imamura A et al. Functional tooth regeneration using a bioengineered tooth unit as a mature organ replacement regenerative therapy. PLoS ONE 2011;6:e21531.
Angelova Volponi A, Kawasaki M, Sharpe PT . Adult human gingival epithelial cells as a source for whole-tooth bioengineering. J Dent Res 2013;92:329–334.
Kitagawa M, Tahara H, Kitagawa S et al. Characterization of established cementoblast-like cell lines from human cementum-lining cells in vitro and in vivo. Bone 2006;39:1035–1042.
Harada H, Mitsuyasu T, Nakamura N et al. Establishment of ameloblastoma cell line, AM-1. J Oral Pathol Med 1998;27:207–212.
Hatano H, Kudo Y, Ogawa I et al. Establishment of mesenchymal cell line derived from human developing odontoma. Oral Dis 2012;18:756–762.
Kaneda T, Miyauchi M, Takekoshi T et al. Characteristics of periodontal ligament subpopulations obtained by sequential enzymatic digestion of rat periodontal ligament. Bone 2006;38:420–426.
Alimperti S, Andreadis ST . CDH2 and CDH11 act as regulators of stem cell fate decisions. Stem Cell Res 2015;14:270–282.
Czopka T, von Holst A, ffrench-Constant C et al. Regulatory mechanisms that mediate tenascin C-dependent inhibition of oligodendrocyte precursor differentiation. J Neurosci 2010;30:12310–12322.
Peles E, Nativ M, Campbell PL et al. The carbonic anhydrase domain of receptor tyrosine phosphatase beta is a functional ligand for the axonal cell recognition molecule contactin. Cell 1995;82:251–260.
Peles E, Schlessinger J, Grumet M . Multi-ligand interactions with receptor-like protein tyrosine phosphatase beta: implications for intercellular signaling. Trends Biochem Sci 1998;23:121–124.
Na J, Baker D, Zhang J et al. Aneuploidy in pluripotent stem cells and implications for cancerous transformation. Protein Cell 2014;5:569–579.
Ben-David U, Benvenisty N . The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer 2011;11:268–277.
Schmitd LB, Bravo-Calderón DM, Soares CT et al. Hyperplastic dental follicle: a case report and literature review. Case Rep Dent 2014;2014:251892.
Foster BL, Popowics TE, Fong HK et al. Advances in defining regulators of cementum development and periodontal regeneration. Curr Top Dev Biol 2007;78:47–126.
Luan X, Ito Y, Diekwisch TGH . Evolution and development of Hertwig's epithelial root sheath. Dev Dyn 2006;235:1167–1180.
Thomas HF . Root formation. Int J Dev Biol 1995;39:231–237.
Zeichner-David M . Regeneration of periodontal tissues: cementogenesis revisited. Periodontol 2000 2006;41:196–217.
Nam H, Kim JH, Kim JW et al. Establishment of Hertwig's epithelial root sheath/epithelial rests of Malassez cell line from human periodontium. Mol Cells 2014;37:562–567.
Philipsen HP, Reichart PA, Slootweg PJ et al. Neoplasms and tumour-like lesions arising from the odontogenic apparatus and maxillofacial skeleton: introduction. In: Barnes L, Eveson JW, Reichart P, (eds). World Health Organization Classification of Tumours. Pathology and Genetics of Head and Neck Tumours. IARC Press: Lyon, France, 2005, pp 285–286.
Lin LM, Huang GT, Rosenberg PA . Proliferation of epithelial cell rests, formation of apical cysts, and regression of apical cysts after periapical wound healing. J Endod 2007;33:908–916.
Acknowledgements
This study was supported by a Grant-in-Aid from the Ministry of Education, Science, and Culture of Japan. Technical assistance was provided from the Support Center for Advanced Medical Sciences (Tokushima University). We thank Mr Hideaki Horikawa (Tokushima University) for technical assistance. The authors also thank Dr Hidemitsu Harada (Iwate Medical University) for providing AM-1 cells and Dr NE Fusenig (German Cancer Research Center) for providing HaCaT cells.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Supplementary information
Rights and permissions
About this article
Cite this article
Tsunematsu, T., Fujiwara, N., Yoshida, M. et al. Human odontogenic epithelial cells derived from epithelial rests of Malassez possess stem cell properties. Lab Invest 96, 1063–1075 (2016). https://doi.org/10.1038/labinvest.2016.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2016.85
This article is cited by
-
Analysis of the cells isolated from epithelial cell rests of Malassez through single-cell limiting dilution
Scientific Reports (2022)
-
Direct reprogramming of epithelial cell rests of malassez into mesenchymal-like cells by epigenetic agents
Scientific Reports (2021)
-
Isolation and characterization of cells derived from human epithelial rests of Malassez
Odontology (2019)