Abstract
Adverse health outcomes in pulmonary fibrosis are associated with extracellular matrix (ECM) accumulation. Although transforming growth factor-β (TGF-β) has been reported to be an important regulator of fibrosis pathogenesis, TGF-β-independent pathways may also be involved. Here, we investigated responses of putative relatively fibrosis-resistant BALB/c mice to transient pulmonary overexpression of oncostatin M (OSM) using an adenovirus vector encoding OSM (AdOSM) and compared responses with the relatively fibrosis-prone C57Bl/6 strain. Interestingly, BALB/c mice showed similar ECM accumulation and collagen 1A1 and 3A1 mRNA elevation to C57Bl/6 mice 7 days after endotracheal administration of AdOSM. TGF-β1 mRNA levels and pSMAD2 signal were not regulated in either strain in total lung extracts. In contrast to C57Bl/6 mice, BALB/c mice lacked eosinophil, Th2 cytokine, and pro-inflammatory cytokine elevation in the broncholveolar space. OSM overexpression induced STAT3 activation and SMAD1/5/8 signaling suppression in lung from both mice strains, which was associated with a downregulation of BMPR2 and BMP ligands, and increased expression of the BMP antagonist gremlin. Although we also observed STAT3 activation and SMAD1/5/8 signaling suppression in mouse lung fibroblast cultures in vitro upon OSM stimulation, immunohistochemistry analyses indicated that the AdOSM-induced pSMAD1/5/8 signal suppression was primarily localized to the airway epithelium. Other gp130 cytokines including IL-6, LIF, CT-1, but not IL-31, also induced STAT3 activation and SMAD1/5/8 signaling suppression in C10 mouse lung epithelial cells and BEAS 2B bronchial epithelial cells, and we found that pharmacological inhibition of STAT3 activation reversed OSM-induced SMAD1/5/8 signaling suppression in vitro. The results demonstrate that OSM induces ECM accumulation in fibrosis-resistant BALB/c mouse lung in the absence of Th2 inflammation or TGF-β signaling, and highlight a dichotomy of STAT3 activation versus SMAD1 suppression in this process.
Similar content being viewed by others
Main
Chronic and excessive remodeling of the extracellular matrix (ECM) is a prominent finding in pulmonary fibrosis.1, 2 Pulmonary fibrosis is a debilitating condition of the lung interstitium characterized by thickening of the alveolar walls, airway epithelial cell hyperplasia, and lung stiffness.1, 2 The number of deaths associated with the subtype idiopathic pulmonary fibrosis (IPF) is similar to certain types of cancer, and the median prognosis for survival once diagnosed with this condition is 2.5–3.5 years.3, 4 The current paradigm in pulmonary fibrosis pathogenesis is that ECM accumulation is driven by epithelial damage and fibroblast proliferation, processes that involve the profibrotic cytokine transforming growth factor-β (TGF-β).5 TGF-β has been shown to induce collagen production in lung fibroblasts,6 and to induce epithelial cell differentiation into α-smooth muscle actin (α-SMA)-positive myofibroblasts through epithelial-to-mesenchymal transition (EMT).7, 8, 9 Pulmonary overexpression of active TGF-β using AdTGF-β induces ECM remodeling in rats and C57Bl/6 mice, but interestingly not in BALB/c mice.10 This mirrors trends seen in the bleomycin model of pulmonary fibrosis, thought to be primarily driven by TGF-β, where C57Bl/6 mice are more sensitive to bleomycin than BALB/c mice.11 Although clear implications of TGF-β are evident in preclinical studies of pulmonary fibrosis, clinical trials targeting TGF-β alone have not yet reported success for this disease. TGF-β-independent mechanisms involved in ECM remodeling may also contribute in certain situations.
Among numerous growth factors, several members of the gp130 cytokine family have been implicated in lung ECM remodeling including IL-11 and oncostatin M (OSM).12, 13 We and others have previously shown that transient pulmonary elevation of OSM in C57Bl/6 mice induced rapid ECM remodeling.13, 14 In cell culture, OSM has been described as a mitogenic, anti-apoptotic factor that stimulates collagen production in human lung fibroblasts.15 Elevated levels of OSM have been detected in the bleomycin model of pulmonary fibrosis as well as the bronchoalveolar lavage fluid (BALF) of IPF patients, suggesting a potential role for OSM in pulmonary fibrosis.14, 16
Cell signaling induced by TGF-β in numerous cell types include the activation of the canonical SMAD2/3 activation pathway.17 In contrast, downstream cell signaling pathways of gp130 cytokines include the janus kinase (JAK) and signal transducer and activator of transcription (STAT) pathway.18, 19 Gp130757F mice with hyperactive STAT3 showed exacerbated fibrosis in response to bleomycin, whereas gp130ΔSTAT mice with hypoactive STAT3 showed attenuated effects upon bleomycin treatment.20 STAT3 activation in human lung fibroblasts has also been shown to induce several morphological features consistent with a myofibroblast phenotype.21 Thus, evidence suggests that STAT3 activation contributes to fibrosis.
Bone morphogenetic proteins (BMPs) are part of the TGF-β superfamily. BMP ligands activate SMAD1/5/8 signaling and several BMP ligands have a protective role upon lung injury. Administration of either recombinant BMP-7 or tilorone, both agonists of the canonical BMP pathway SMAD1/5/8, showed therapeutic effects in a mouse model of silica-induced pulmonary fibrosis.22, 23 The BMP antagonist gremlin has been associated with alveolar epithelial cell (AEC) activation with proliferation and apoptosis, as well as IPF.24, 25 However, the involvement of the BMP pathway during lung injury is not well characterized in other experimental models.
In this study, we examined and compared the effects of transient pulmonary overexpression of OSM in BALB/c and C57Bl/6 mice. Contrary to observations using the bleomycin and AdTGF-β models, OSM overexpression induced ECM accumulation and collagen 3a1 mRNA levels in both mouse strains, and this mirrored responses of mouse lung fibroblast (MLF) cultures to OSM in vitro. In addition, the local lung inflammatory response of BALB/c mice did not show a Th2-skewed immune response as predicted by the paradigm of Th1/Th2 responses in these two strains of mice. Furthermore, OSM did not induce canonical TGF-β/SMAD2/3 pathways in vivo, but did result in a dichotomy of STAT3 and SMAD1 regulation that was associated with a downregulation of BMPR2 and BMP ligands as well as increased gremlin expression. Although regulation of these pathways were observed in MLF cultures in vitro upon OSM stimulation, pSMAD1/5/8 signal suppression was localized to airway epithelial cells, as determined by immunohistochemistry. Using C10 mouse lung epithelial cells and BEAS 2B bronchial epithelial cells, we observed that other gp130 cytokines IL-6, LIF, and CT-1 induced STAT3 activation and SMAD1/5/8 signaling suppression, and that inhibition of the STAT3 pathway using a small molecule inhibitor Stattic reversed OSM-induced pSMAD1/5/8 suppression.
MATERIALS AND METHODS
Mice
Female BALB/c and C57Bl/6 mice (6–8 weeks of age) were purchased from Charles River (Ottawa, ON, Canada). Approval for all in vivo experimentation described in this study was obtained from the McMaster Animal Research Ethics Board (Hamilton, ON, Canada).
OSM Overexpression in Mice
As previously described,13 mice (n≥4/group) were endotracheally administered 5 × 107 PFUs of either the replication-deficient adenovirus encoding OSM (AdOSM) or control vector AdDl70. Samples were collected after 5, 7, 14, and 28 days.
Collection of BALF and Lung Samples
BALF was collected as previously described for cytokine and inflammatory cell analyses.26 Total cell counts were measured using a hemocytometer, and cell differentials were conducted by staining cytocentrifuge smears (Hema-3 Stain, Fisher Scientific) and subsequently identifying cells on the basis of standard hemocytologic criteria for macrophages/monocytes, eosinophils, neutrophils, and lymphocytes. The left lung was perfused with 10% formalin and prepared for histology and immunohistochemistry analyses. The right lung was snap frozen, stored, and prepared for mRNA and protein analysis as previously described.13 BALF sample levels of cytokine were quantified for KC, IL-1β, TNF-α, VEGF, eotaxin-2, IL-4, IL-5, and IL-10 using LUMINEX bead assays (Luminex, Toronto, ON, Canada) following the manufacturer’s recommendations.
Histology and Immunohistochemistry
Lung sections (3 μm) were cut from paraffin blocks and stained with H&E to assess pathology, and picrosirius red (PSR) to assess collagen content. To quantify lung interstitial collagen, four individuals blinded to the experimental design rated the extent of interstitial PSR staining (0–8 score, with 8 being most severe) evident in photomicrographs of PSR-stained slides examined under polarized light (>8 photomicrographs per animal to cover entire lung section). The extent of staining specifically within the lung interstitium was assessed, as it has been previously shown that AdOSM induces collagen in lung interstitium but not the major airways.13 Immunohistochemistry for α-SMA and phospho-SMAD1/5/8 (Millipore) was performed following the manufacturer’s protocol that included diluting the primary antibodies 1:100 and using the Dako EnVision+ System-HRP Anti-Rabbit detection system (Dako, Oakville, ON, Canada).
Lung Homogenization for Protein and RNA Isolation
Crushed lung powder in either Trizol or RIPA buffer was homogenized (Brinkmann Homogenizer Polytron PT 3000). RNA was isolated from Trizol following the manufacturer’s protocol. For protein isolation, homogenized tissue was spun for 30 min at 2000 r.p.m., and the supernatant was collected. RNA and protein were both stored at −80 °C until further use.
Cell Culture
BALB/c and C57Bl/6 MLF cultures were isolated from whole lung as previously described.27 BEAS 2B human bronchial epithelial cells (<passage 8) and C10 mouse lung epithelial cells (<passage 8) were received as a gift from the laboratory of Dr Mark Larché (McMaster University) and Dr Chris Migliaccio (University of Montana), respectively. Cells were treated with 5 ng/ml OSM, 50 ng/ml BMP-2, 5 ng/ml TGF-β1, 5 ng/ml IL-6, 5 ng/ml LIF, 5 ng/ml CT-1, 5 ng/ml IL-11, or 5 ng/ml IL-31 (all from R&D, Minneapolis, MN, USA), and collected for mRNA and protein analyses using standard protocols. To determine the effects of inhibiting STAT3 activation on SMAD1/5/8 signaling, BEAS-2B cells were pretreated with a small molecule inhibitor Stattic for 1 h before cytokine treatment.28
RNA Analysis by qRT-PCR (Taqman)
RNA samples were quantified using a NanoVue Spectrophotometer (GE Healthcare, Little Chalfont, UK). Genomic DNA was removed using Ambion DNA-free kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s directions. cDNA was synthesized from RNA using the manufacturer’s instructions, and probed by q-RT-PCR for: collagen 1a1, collagen 3a1, TIMP-1, MMP-13, IL-4, IL-5, eotaxin-1, eotaxin-2, TGF-β1, BMP-2, BMP-4, gremlin, and Id1 (all from Life Technologies). mRNA expression for each sample was normalized using the ΔΔCT method against 18S expression.13 The use of 18S as a reference gene was validated among a panel of candidates (18S, Actb, B2M, Ccna2, Gapdh, Gusb, Hrpt1, Ipo8, Pgk1, Ppia, Tbp, Ubc, and Ywhaz) from RNA derived from the lungs of mice treated with either AdOSM or AdDl70 for 7 days. 18S showed the highest stability and was thus used to normalize data in all subsequent analysis.
Protein Analysis by Immunoblots
Protein samples were quantified by Bio-Rad protein assay (Bio-Rad Laboratories, Mississauga, ON, Canada), separated by SDS-PAGE, and subsequently transferred onto nitrocellulose membranes using standard methods. Blots were blocked in 5% milk diluted in 1 × TBS with 0.15% Tween, and probed using the following antibodies: Actin I-19 (Santa Cruz Biotechnology, Dallas, TX, USA), phospho-SMAD1/5/8 (Millipore, Billerica, MA, USA), phosho-STAT3, phospho-SMAD1/5, SMAD1, phospho-SMAD2, SMAD2, and BMPR2 (all from Cell Signal, Danvers, MA, USA). Finally, corresponding secondary antibodies and ECL2 (Thermo Scientific, Ottawa, ON, Canada) were used to develop the blots. Densitometry of band intensity was completed using ImageJ software (NIH, Washington DC, USA), normalized to actin, SMAD1, or SMAD2 (as indicated), and compared relative to untreated/control samples.
Statistical Analysis
Statistical tests, including unpaired t-test and one-way ANOVA, were conducted using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). All error bars indicate the mean standard error. A minimum of three, but usually four or five, mice were used per treatment group in each experiment and the experiments were repeated at least once. A P-value of <0.05 was considered statistically significant.
RESULTS
AdOSM Induces Lung ECM Accumulation in Both BALB/c and C57Bl/6 Mice
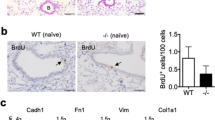
In previous work, we described responses of C57Bl/6 mice to endotracheal administration of AdOSM that included ECM accumulation in the lung interstitium as well as a Th2-type response in the lung by 7 days.13 As BALB/c mice are relatively protected in other experimental models of pulmonary fibrosis, we directly compared responses in these two strains with AdOSM treatment. Histopathological examination revealed that AdOSM but not AdDl70 (each at 5 × 107 PFUs) induced inflammatory cell accumulation and thicker alveolar walls in BALB/c mice at day 7 (both, Figure 1a). These changes to the alveoli architecture were more pronounced at day 14 but attenuated at day 28 (Figure 1a). Quantification of interstitial PSR staining under polarized light determined that AdOSM induced interstitial collagen accumulation in both BALB/c and C57Bl/6 mice (Figures 1b and c). Furthermore, OSM overexpression resulted in increased α-SMA staining in the lung interstitial of BALB/c mice, whereas α-SMA was detected only around blood vessels and large airways in AdDl70-treated mice (Figure 1d).
Extracellular matrix deposition upon administration of AdOSM vector. BALB/c and C57BL/6 mice were endotracheally administered AdDl70 or AdOSM vectors (5 × 107 PFUs; n≥4 per group) and culled 7, 14, or 28 days later. Lung sections were prepared as described in the Materials and Methods and stained for H&E (a) and PSR (b). (c) Quantification of extent of interstitial PSR staining was completed using modified Ashcroft scoring as described in the Materials and Methods and results shown represent the mean±s.e.m. Statistical significance is indicated where P<0.05 (*P<0.05, **P<0.01, ***P<0.001). (d) Lung slices from BALB/c mice were stained for α-SMA by IHC, and insets show negative control. Arrows indicate positive SMA staining. The scale bar for all photomicrographs represents a length of 100 μm.
BALB/c Versus C57Bl/6 Inflammatory Response to AdOSM
To characterize inflammatory cell infiltrates into the lung, we identified BALF cells using cytocentrifuge smears. AdOSM-treated BALB/c mice showed increased percentages of neutrophils and lymphocytes (Figure 2a) relative to naive and AdDl70-treated BALB/c mice. On the other hand, eosinophils were not detected in the BALF of AdOSM-treated BALB/c mice (Figure 2a), in contrast to previously published responses in C57Bl/6 mice using the same AdOSM vector.13, 26 Cytokine and chemokine levels were also measured in BALF samples at day 7. KC levels were elevated in AdOSM-treated mice of both strains (Figure 2b). On the other hand, eotaxin-2 levels were below the limit of detection in BALB/c mice upon any treatment, but markedly elevated in AdOSM-treated C57Bl/6 mice. Furthermore, no detectable levels were evident for the Th2 cytokines IL-4, IL-5, and IL-10 or the inflammatory cytokines IL-1β and TNF-α in the BALF of AdOSM-treated BALB/c mice. In parallel experiments, these cytokines were markedly elevated in AdOSM-treated C57Bl/6 mice (Figure 2b). In contrast to the pro-inflammatory and Th2 cytokines, VEGF levels were elevated in AdOSM-treated BALB/c mice and below the limit of detection in C57Bl/6 mice.
Inflammatory cells and cytokine levels in BALF upon administration of AdOSM vector. (a) BALB/c mice were left untreated (naive) or endotracheally administered AdDl70 or AdOSM vectors (5 × 107 PFUs; n≥3) and culled 7 days later. Percent of macrophages, lymphocytes, eosinophils, and neutrophils were determined in the BALF of BALB/c mice. (b) Protein levels of IL-6, VEGF, TNF-α, IL-1β, eotaxin-2, IL-4, IL-5, and IL-10 were determined in both BALB/c and C57Bl/6 mice by ELISA, where each point represents a separate mouse. The results represent the mean±s.e.m., and statistical significance is indicated where P<0.05 (*P<0.05, **P<0.01, ***P<0.001).
AdOSM Induces mRNA for Collagen But Not Th2 Cytokines in BALB/c Mice In Vivo
The mRNA analyses using day 7 lung homogenates showed significant elevation of collagen 3a1 steady-state mRNA levels in both mice strains upon OSM overexpression (Figure 3a). TGF-β1 mRNA levels tended to decrease upon AdOSM but these changes were not statistically significant in either strain. The mRNA levels of chemotactic factors for eosinophils was also determined at day 7, and showed in BALB/c mice that AdOSM induced eotaxin-1 but not eotaxin-2 gene expression. In contrast, AdOSM-treated C57Bl/6 mice showed markedly increased levels of both eotaxin-1 and eotaxin-2 mRNA. Steady-state mRNA levels of IL-4 and IL-5 in the lung were assessed to verify protein levels in the BALF and were both found at elevated levels in AdOSM-treated C57Bl/6 but not BALB/c mice. mRNA analysis and comparison between BALB/c and C57Bl/6 mice was also completed at an earlier time point using day 5 lung homogenates. In contrast to day 7 results, only AdOSM-treated BALB/c mice showed significant increases in collagen 3a1 mRNA levels. Similar trends in collagen 1a1, TIMP-1, and MMP-13 mRNA levels were observed (Figure 3b).
Total lung mRNA in mice upon administration of AdOSM vector. BALB/c and C57BL/6 mice were endotracheally administered AdDl70 or AdOSM vectors (5 × 107 PFUs; n≥4) and culled 5 or 7 days later. (a) Steady-state mRNA levels of collagen 3A1, TGF-β1, eotaxin-1, eotaxin-2, IL-4, and IL-5 were assessed from total lung at day 7. (b) Lung samples were assessed for steady-state mRNA levels of collagen 3a1, collagen 1a1, TIMP-1, and MMP-13 at 5 days after AdDl70 or AdOSM (5 × 107 PFUs) administration. Steady-state mRNA levels were corrected to 18S levels and expressed as fold changes relative to AdDl70-treated mice. White bars represent BALB/c mice whereas black bars represent C57Bl/6 mice. The results represent the mean±s.e.m., and statistical significance is indicated where P<0.05 (*P<0.05, **P<0.01, ***P<0.001).
AdOSM Dysregulates STAT3 and SMAD1 Signaling in Lung Homogenates
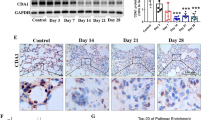
To examine intracellular signaling pathways engaged by AdOSM in vivo, lung homogenates (day 7) were analyzed using immunoblots and quantitative densitometry for activation of STAT and SMAD pathways. All AdOSM-treated BALB/c and C57Bl/6 mouse lung homogenates showed increased levels of pSTAT3 signal relative to AdDl70-treated mice (Figure 4). In contrast, OSM overexpression caused a decrease in pSMAD1/5 signal in lung homogenates from both strains (Figure 4). This observation was verified using a second antibody targeting pSMAD1/5/8 (Figure 4). The decrease in pSMAD1/5 and pSMAD1/5/8 signal was associated with a decrease in BMPR2 signal (Figure 4). No alterations in SMAD1, pSMAD2, or SMAD2 signal were detected in either AdOSM-treated BALB/c or C57Bl/6 mice at this time point (Figure 4).
STAT3 and SMAD signaling in mouse lung upon administration of AdOSM vector. BALB/c and C57BL/6 mice were endotracheally administered AdDl70 or AdOSM vectors (5 × 107 PFUs; n≥4 per group) and culled 7 days later. (a) Lung homogenates were prepared as described in the Materials and Methods for immunoblots, and probed for pSTAT3, pSMAD1/5, pSMAD1/5/8, SMAD1, BMPR2, pSMAD2, SMAD2, and Actin as indicated. Each lane represents a separate mouse. (b) Quantification of band signal strength was completed using ImageJ as described in the Materials and Methods section (white bars: BALB/c mice; black bars: C57Bl/6 mice). pSTAT3, BMPR2 were normalized to actin; pSMAD1/5 and pSMAD1/5/8 were normalized to SMAD1; pSMAD2 was normalized to SMAD2. The results represent the mean±s.e.m., and statistical significance is indicated where P<0.05 (*P<0.05, **P<0.01, ***P<0.001).
OSM Responses in Mouse Lung Fibroblasts In Vitro
As fibroblasts are an integral cell population in the regulation of the ECM, responses of lung fibroblasts derived from BALB/c and C57Bl/6 mice were examined. Upon stimulation with OSM for 24 h, both BALB/c and C57Bl/6 MLF cultures showed elevation of collagen 3a1 mRNA (Figure 5a). TGF-β1 also induced collagen 3a1 mRNA, but to a lower extent. OSM did not induce collagen 1a1 mRNA whereas TGF-β1 induced levels, although marginally. BMP-2 was also tested as it can regulate collagen in other cell types, but was found to have no stimulatory effect on collagen 3a1 or 1a1 mRNA. However, BMP-2 did upregulate a typical downstream BMP-2 target Id1 mRNA in C57Bl/6 MLF cultures, whereas Id1 mRNA was suppressed by TGF-β1 stimulation.
Effects of BALB/c and C57Bl/6 MLF cultures in vitro upon OSM stimulation. BALB/c and C57Bl/6 MLF explants were cultured as described in the Materials and Methods and stimulated with 5 ng/ml OSM, 50 ng/ml BMP-2, or 5 ng/ml TGF-β1. (a) mRNA was isolated after 24 h of stimulation, converted to cDNA, and probed for collagen 3a1, collagen 1a1, and Id1 as indicated, corrected to 18S levels, and expressed as fold change from unstimulated (control) cells. (b) BALB/c MLF cell lysates were collected after 72 h of stimulation, prepared for immunoblot, and probed for pSTAT3, pSMAD1/5/8, SMAD1, BMPR2, pSMAD2, SMAD2, and actin. (c) Quantification of bands was completed using ImageJ. pSTAT3 and BMPR2 were normalized to actin; pSMAD1/5/8 was normalized to SMAD1; pSMAD2 was normalized to SMAD2. White bars and black bars represent BALB/c and C57Bl/6 MLF cultures, respectively.
STAT3 and SMAD1/5/8 cell signaling pathways were also investigated in vitro in MLF cultures to compare with the findings observed in vivo. OSM induced pSTAT3 signal, suppressed pSMAD1/5/8 signal, and did not regulate either the BMPR2 or pSMAD2 signal in either BALB/c (Figure 5b) or C57Bl/6 (data not shown) MLF in vitro, whereas BMP-2 and TGF-β1 induced pSMAD1/5/8 and pSMAD2 signals, respectively.
SMAD1/5/8 Signaling Regulation in Airway Epithelial Cells
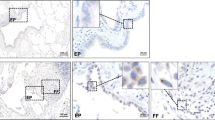
IHC was performed to identify which cells in the lung showed modulation of SMAD1/5/8 signaling. Figure 6a includes representative photomicrographs of staining that showed consistent results. Interestingly, the AdOSM but not the AdDl70 vector caused a marked suppression of pSMAD1/5/8 signal levels in the airway epithelial cells of both BALB/c and C57Bl/6 mice (Figure 6a). This was consistent in multiple fields per mouse lung section examined, and observed at both day 2 and day 7 (day 2 data not shown). Thus, we next examined a mouse lung epithelial cell line (C10 mouse lung epithelial cells) and observed that OSM stimulation caused a decrease in pSMAD1/5/8 signal whereas pSTAT3 signal was elevated after 72 h of stimulation (Figure 6b). Similar findings were observed using the pSMAD1/5 antibody (data not shown). As positive controls for SMAD1 and SMAD2 activation, BMP-2 and TGF-β1 increased pSMAD1/5/8 and pSMAD2 signal, respectively (Figure 6b). In addition, it was shown that OSM was unable to regulate BMPR2 in C10 mouse lung epithelial cells (Figure 6b). As there is overlap in signaling pathways activated by the family of gp130 cytokines, we tested whether this STAT3/SMAD1 imbalance could also be induced by IL-6, LIF, CT-1, or IL-31 in C10 mouse lung epithelial cells. After 72 h of incubation, it was shown that all cytokines except IL-31 were able to induce STAT3 activation and SMAD1/5/8 signaling suppression (Figure 6c).
STAT3 and SMAD1 signaling in epithelial cells upon OSM stimulation. (a) BALB/c and C57BL/6 mice were left untreated or endotracheally administered AdDl70 or AdOSM vectors (5 × 107 PFUs) and culled 7 days later (as in Figure 1). Lung samples were stained for pSMAD1/5/8 by IHC as described in the Materials and Methods. The scale bar for the photomicrographs represents a length of 150 μm, and insets show negative controls for the IHC staining. (b) C10 mouse lung epithelial cells in culture were untreated or stimulated with 5 ng/ml OSM, 50 ng/ml BMP-2, or 5 ng/ml TGF-β1 for 72 h. (c) C10 mouse lung epithelial cells were untreated or treated with 5 ng/ml of OSM, IL-6, LIF, CT-1, or IL-31 for 72 h. In both experiments, cell lysates were collected, prepared for immunoblot, and probed for pSTAT3, pSMAD1/5/8, SMAD1, BMPR2, pSMAD2, SMAD2, or actin as indicated. Quantification of bands was completed using ImageJ. pSTAT3 and BMPR2 were normalized to actin; pSMAD1/5/8 was normalized to SMAD1; pSMAD2 was normalized to SMAD2.
STAT3 Inhibition Reverses OSM-Induced pSMAD1/5/8 Suppression
Stattic is a small molecular inhibitor of STAT3 that prevents the activation, dimerization, and translocation of the STAT3 complex into the nucleus.28 Although it has been previously shown to be effective in inhibiting STAT3 activation after 1 h of incubation, we were unable to incubate C10 mouse lung epithelial cells for 72 h with Stattic without inducing significant cell death. Thus, we went on to utilize BEAS-2B bronchial epithelial cells, as we were able to show STAT3 activation and SMAD1/5/8 signaling suppression upon 1 h of stimulation with OSM or IL-6, but neither LIF nor IL-11 (Figures 7a and b). To determine whether OSM-induced STAT3 activation is required for SMAD1/5/8 signaling suppression, BEAS-2B cells were pretreated with increasing concentrations of Stattic for 1 h before stimulation with 5 ng/ml OSM for another hour. Pretreatment of BEAS-2B cells with 5 μM of Stattic resulted in a marked decrease in pSTAT3 signal and increase in pSMAD1/5/8 signal (Figures 7c and d).
STAT3 activation correlation with SMAD1/5/8 signaling suppression. (a) BEAS 2B bronchial epithelial cells in culture were untreated or stimulated with 5 ng/ml OSM, IL-6, LIF, or IL-11 for 1 h. Cell lysates were collected, prepared for immunoblot, and probed for pSTAT3, pSMAD1/5/8, SMAD1, and actin as indicated. (b) Quantification of bands was completed using ImageJ. (c) Human BEAS-2B cells in culture were pretreated with the indicated micromolar concentrations of Stattic for 1 h. Subsequently, cells were stimulated with 5 ng/ml OSM for 1 h. Cell lysates were collected, prepared for immunoblot, and probed for pSTAT3, pSMAD1/5/8, SMAD1, and actin as indicated. (d) Quantification of bands was completed using ImageJ.
AdOSM Alters Expression of BMP-Associated Ligands
As SMAD1 signaling was reduced by OSM, we next examined typical SMAD1 signaling modulators including BMP-2, BMP-4, and gremlin. Lung homogenates from mice at 7 days after AdOSM administration showed a downregulation of BMP-2 and BMP-4 mRNA in both strains of mice (Figure 8). This contrasted with increased gremlin mRNA levels in both strains of mice in response to AdOSM (Figure 8).
Total lung mRNA levels for BMP ligands in mice upon administration of AdOSM vector. BALB/c and C57BL/6 mice were endotracheally administered AdDl70 or AdOSM vectors (5 × 107 PFUs; n≥4 per group) and culled 7 days later. Lung samples (as in Figure 3a) were assessed for steady-state mRNA levels of BMP-2, BMP-4, and gremlin. The results represent the mean±s.e.m., and statistical significance is indicated where P<0.05 (*P<0.05, **P<0.01, ***P<0.001).
DISCUSSION
Our results show that transient pulmonary overexpression of OSM induces comparable ECM deposition in both the fibrosis-resistant BALB/c and fibrosis-prone C57Bl/6 mice after 7 days. Furthermore, in contrast to C57Bl/6 mice, BALB/c mice lacked eosinophils and elevation of the Th2 cytokines IL-4, IL-5, and IL-10, as well as TNF-α and IL-1β in bronchoalveolar spaces. This also suggests that eosinophils or Th2 immune mediators are not required for ECM accumulation in these models. In both strains, there was marked STAT3 activation but SMAD1 signaling inhibition, with the latter effect being localized to the airway epithelial cells. Other members of the gp130 cytokine family also induced this dichotomy of STAT3/SMAD1 signaling and inhibition of STAT3 reversed SMAD1/5/8 signaling suppression. Lastly, the suppression of SMAD1/5/8 signaling was associated with a downregulation at the mRNA level of BMP ligands BMP-2 and -4, and an upregulation of the BMP antagonist gremlin.
OSM overexpression induced ECM as measured by PSR collagen staining (Figure 1c) that has previously been shown to correlate with increased interstitial collagen deposition.13 An examination of mRNA levels from lung homogenates showed that OSM overexpression induced ECM remodeling genes. Collagen 3a1 mRNA levels were significantly elevated in both strains by day 7 (Figure 3a), but only so in BALB/c mice at day 5 (Figure 3b). Similarly, collagen 1a1, TIMP-1, and MMP-13 mRNA levels were significantly elevated in AdOSM-treated BALB/c but not C57Bl/6 mice at day 5 (Figure 3b), and together these data suggest that AdOSM-induced ECM remodeling occurs more rapidly in BALB/c than in C57Bl/6 mice, despite the absence of eosinophils, Th2-type cytokines, and pro-inflammatory cytokines in the bronchoalveolar spaces.
The ratio between type III and type I collagen during wound repair has received considerable attention in recent years. Type III collagen is produced during the early reparative process and contributes to a provisional matrix, whereas type I collagen emerges at later stages of wound repair.29, 30 Intriguingly, we show here that OSM was a more potent inducer of collagen 3a1 than TGF-β1 in both BALB/c and C57Bl/6 MLF in vitro (Figure 5a). However, in contrast to TGF-β1, OSM was unable to induce collagen 1a1 expression in either MLF culture system. Our data may implicate OSM in early tissue remodeling mechanisms. Currently, it is unknown how OSM regulates collagen 3a1 differentially from collagen 1a1, and this may be because of differences in the promoter regions of these two collagen genes. The collagen 1a1 expression that we observed in whole lung RNA could be because of other cell types or induced by other factors in vivo. However, TGF-β1-mediated effects appear not to participate in the AdOSM system as levels of TGF-β1 mRNA (Figure 3a) and pSMAD2 signal (Figure 4) were unregulated in lung homogenates at 7 days after administration of AdOSM. Although pSMAD3 signaling was not examined in this study, it has been previously shown that AdOSM induces interstitial ECM remodeling in SMAD3 knockout mice that was comparable to wild-type C57Bl/6 mice.20 Alternatively, collagen 1a1 expression may be regulated at a different time point by OSM in MLF cultures in vitro, although similar trends were observed after 6 and 72 h of stimulation (both unpublished data generated by Steven Wong).
The regulation of ECM remodeling in various tissues may involve cell signaling pathways including STAT3. For instance, STAT3 activation through IL-6 transsignaling in hypertrophic scar fibroblasts was associated with fibronectin and type I collagen induction, although that study did not specifically address collagen 3a1.31 We observed in this study that AdOSM-induced ECM accumulation correlated with STAT3 activation (Figure 4), and that OSM activated STAT3 in vitro in both MLF cultures and C10 mouse lung epithelial cells (Figures 5b and 6b). A recent work by O’Donoghue et al20 showed that transgenic mice with hyperactive STAT3 were more prone to bleomycin-induced lung fibrosis.20 The function of STAT3 activation appears complex, as different cell types behave differently upon STAT3 activation. For example, STAT3 is required for the differentiation of fibroblasts into myofibroblasts;21 however, other studies showed that in a population of pSTAT3+ fibroblasts from IPF patients, inhibition of STAT3 signaling increased α-SMA levels.32 STAT3 signaling in the airway epithelial cells is also thought to restore cell shape and is required for repair of the bronchiolar epithelium after injury,33 but other studies show that STAT3 activation is required for differentiation of normal human squamous epithelial cells. The complex role of STAT3 activation therefore warrants further investigation.
In addition to STAT3 activation, we also showed that AdOSM-induced ECM accumulation correlated with SMAD1 suppression and, to our knowledge, this is the first study to show a dichotomy between these pathways. In fact, our data in C10 mouse lung epithelial cells showing that OSM, IL-6, LIF, CT-1, but not IL-31, are able to induce this STAT3/SMAD1 signaling dichotomy (Figure 7a) suggest that STAT3 activation may be linked to SMAD1/5/8 signaling suppression. Furthermore, we have evidence linking STAT3 activation to SMAD1/5/8 signaling suppression as inhibiting the STAT3 pathway using the pharmacological inhibitor Stattic reversed OSM-induced SMAD1/5/8 inhibition (Figure 7b). Like STAT3, SMAD1 regulation has more recently been implicated in fibrosis pathogenesis. SMAD1/5/8 signaling suppression has been observed in the silica model of pulmonary fibrosis in mice,22, 23 and overexpression of the BMP signaling antagonist gremlin in rat lung causes ECM deposition.24 The SMAD1 pathway is thought to have a homeostatic role in the maintenance of lung tissue, as the pathway is constitutively activated even in healthy airway epithelium.34 Thus, the BMP–SMAD1 signaling axis is believed to have an antifibrotic function in the lung,22, 25 and based on this suggestion SMAD1 signaling suppression would exacerbate fibrosis. In addition to STAT3 activation, the suppression that we observed in SMAD1 signaling by OSM both in vivo and in vitro could also be explained by a downregulation of BMPR2 (Figure 4) or BMP-2/4 ligands (Figure 8), or upregulation of the BMP antagonist gremlin (Figure 8), or combinations thereof. BMP ligands have been shown to inhibit TGF-β-induced collagen expression in myofibroblasts,35, 36 but it is uncertain whether BMP ligands can similarly inhibit OSM-induced collagen expression. This would be subject to further study, as would the mechanism of a direct cause and effect relationship between OSM-induced SMAD1 pathway suppression and ECM deposition.
The absence of eosinophil infiltration into the lungs of BALB/c mice upon OSM overexpression is in contrast to responses in C57Bl/6 mice.13, 14 The lack of eosinophils was associated with a lack of eotaxin-2 levels in AdOSM-treated BALB/c mice (Figure 2b) despite elevated eotaxin-1 levels. Others have shown that eosinophil infiltration into the alveolar space is highly dependent on both eotaxin-1 and eotaxin-2 in a mouse model of ovalbumin-induced airway inflammation,37 and in response to IL-13-induced airway eosinophilia.38 Our data showing a lack of eotaxin-2 (both mRNA and protein levels) in BALB/c mice in response to OSM may explain the strain difference in eosinophil infiltration. Alternatively, IL-4 and IL-5, both of which showed significant elevation (both mRNA and protein levels) in C57Bl/6 but not BALB/c mice, may be responsible for eosinophil recruitment as eosinophil recruitment has been shown to be dependent on these cytokines.39 However, our data in BALB/c mice suggest that eosinophils or Th2 cytokines do not play an integral role in OSM-induced ECM accumulation, as comparable increases in PSR staining were observed in BALB/c and C57Bl/6 mice despite differences in eosinophil infiltration.
The induction of ECM accumulation and collagen mRNA levels in BALB/c mice are intriguing, in light of the published work showing a resistance of this strain to bleomycin or TGF-β-induced fibrosis.10, 11 This further supports TGF-β-independent pathways induced by OSM in vivo in mouse lungs. Historically, BALB/c mice have displayed more of a Th2-skewed response than C57Bl/6 mice in models of airway allergic diseases.40 Previous work (confirmed in Figure 2b) has shown that OSM overexpression induces Th2 cytokines as well as TNF-α and IL-1β in C57Bl/6 mice.13, 26 However, such responses were not detectable in BALB/c mice in the direct comparison studies completed here. This suggests that OSM can induce ECM accumulation without pro-inflammatory cytokine elevation in BALB/c mice, and this may have relevance in human disease in the context of the recent de-emphasis of the role of classic inflammatory mediators in IPF pathogenesis.41, 42 In addition, IL-4 and IL-5 were not regulated at the protein or mRNA level, indicating a lack of Th2-skewed cytokine response in BALB/c mice in contrast to expectations.13 It is unclear why there is such a differential gene response to the same cytokine and this may be related to differences in strain responses to adenovirus vector, or that OSM overexpression may engage feedback mechanisms differently in these two strains. Despite the differences in inflammatory milieu between the two strains, STAT3 activation, SMAD1 deactivation (with no pSMAD2 modulation), and associated ECM remodeling is similar upon transient pulmonary overexpression of OSM, indicating that pathways to fibrogenesis that are independent of TGF-β or Th2 cytokine skewing can be driven by OSM in both BALB/c and C57Bl/6 mice.
Collectively, these findings provide insight into TGF-β-independent mechanisms of ECM remodeling within the lung. Lung fibroblasts can directly contribute to ECM accumulation, and epithelial cells can initiate wound repair mechanisms. The epithelium lining the airways and the alveoli play an important role as an effective physical barrier. Proper epithelial–mesenchymal cell interactions are critical for maintaining normal tissue function, as damage to the lung epithelium upon exposure to agents such as asbestos, silica, or bleomycin induces morphological changes that disrupt the homeostasis between the epithelium and the underlying mesenchymal cells.43, 44 These changes to the epithelium may lead to EMT and/or the recruitment, activation, and proliferation of lung fibroblasts that leads to subsequent ECM accumulation. In our model, we observed marked STAT3 activation and SMAD1/5/8 signaling suppression in epithelial cells in vivo upon OSM overexpression and further work is required to determine whether OSM can induce EMT through a STAT3/SMAD1 signaling dichotomy. OSM has been reported to induce EMT of kidney tubular epithelial cells.45, 46 In addition to regulating epithelial cells, OSM acts directly on lung fibroblasts to induce ECM accumulation15 and the collagen 3A1 induction that we observed may also require STAT3 and/or SMAD1/5/8 signaling modulation. Although pro-inflammatory cytokines, Th2 cytokines, and eosinophils are elevated in AdOSM-treated C57Bl/6 mice, the lack of these in AdOSM-treated BALB/c mice downplays their role in inducing ECM accumulation during wound repair. Overall, these findings suggest a unique role for OSM in inducing ECM accumulation in vivo in both fibrosis-resistant BALB/c and fibrosis-prone C57Bl/6 mice and suggest that targeting this molecule may be effective in certain pulmonary disorders.
References
Fernandez IE, Eickelberg O . New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet 2012;380:680–688.
Todd NW, Luzina IG, Atamas SP . Molecular and cellular mechanisms of pulmonary fibrosis. Fibrogenesis Tissue Repair 2012;5:11.
Meltzer EB, Noble PW . Idiopathic pulmonary fibrosis. Orphanet J Rare Dis 2008;3:8.
Ley B, Collard HR, King TE . Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2011;183:431–440.
King TE, Pardo A, Selman M . Idiopathic pulmonary fibrosis. Lancet 2011;378:1949–1961.
Raghu G, Masta S, Meyers D et al. Collagen synthesis by normal and fibrotic human lung fibroblasts and the effect of transforming growth factor-beta. Am Rev Respir Dis 1989;140:95–100.
Willis BC, Liebler JM, Luby-Phelps K et al. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332.
Chapman HA . Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol 2011;73:413–435.
Cabrera-Benítez NE, Parotto M, Post M et al. Mechanical stress induces lung fibrosis by epithelial-mesenchymal transition. Crit Care Med 2012;40:510–517.
Kolb M, Bonniaud P, Galt T et al. Differences in the fibrogenic response after transfer of active transforming growth factor-beta1 gene to lungs of “fibrosis-prone” and “fibrosis-resistant” mouse strains. Am J Respir Cell Mol Biol 2002;27:141–150.
Kim JH, Kim HY, Kim S et al. Natural killer T (NKT) cells attenuate bleomycin-induced pulmonary fibrosis by producing interferon-gamma. Am J Pathol 2005;167:1231–1241.
Knight DA, Ernst M, Anderson GP et al. The role of gp130/IL-6 cytokines in the development of pulmonary fibrosis: critical determinants of disease susceptibility and progression? Pharmacol Ther 2003;99:327–338.
Fritz DK, Kerr C, Fattouh R et al. A mouse model of airway disease: oncostatin M-induced pulmonary eosinophilia, goblet cell hyperplasia, and airway hyperresponsiveness are STAT6 dependent, and interstitial pulmonary fibrosis is STAT6 independent. J Immunol 2011;186:1107–1118.
Mozaffarian A, Brewer AW, Trueblood ES et al. Mechanisms of oncostatin M-induced pulmonary inflammation and fibrosis. J Immunol 2008;181:7243–7253.
Scaffidi AK, Mutsaers SE, Moodley YP et al. Oncostatin M stimulates proliferation, induces collagen production and inhibits apoptosis of human lung fibroblasts. Br J Pharmacol 2002;136:793–801.
Leach HG, Chrobak I, Han R et al. Endothelial cells recruit macrophages and contribute to a fibrotic milieu in Bleomycin lung injury. Am J Respir Cell Mol Biol 2013;49:1093–1101.
Moustakas A, Souchelnytskyi S, Heldin CH . Smad regulation in TGF-beta signal transduction. J Cell Sci 2001;114 (Pt 24):4359–4369.
Carbia-Nagashima A, Arzt E . Intracellular proteins and mechanisms involved in the control of gp130/JAK/STAT cytokine signaling. IUBMB Life 2004;56:83–88.
Fujio Y, Maeda M, Mohri T et al. Glycoprotein 130 cytokine signal as a therapeutic target against cardiovascular diseases. J Pharmacol Sci 2011;117:213–222.
O’Donoghue RJJ, Knight DA, Richards CD et al. Genetic partitioning of interleukin-6 signalling in mice dissociates Stat3 from Smad3-mediated lung fibrosis. EMBO Mol Med 2012;4:939–951.
Nagahama KY, Togo S, Holz O et al. Oncostatin M modulates fibroblast function via STAT3. Am J Respir Cell Mol Biol 2013;49:582–591.
Yang G, Zhu Z, Wang Y et al. Bone morphogenetic protein-7 inhibits silica-induced pulmonary fibrosis in rats. Toxicol Lett 2013;220:103–108.
Leppäranta O, Tikkanen JM, Bespalov MM et al. Bone morphogenetic protein-inducer tilorone identified by high-throughput screening is antifibrotic in vivo. Am J Respir Cell Mol Biol 2013;48:448–455.
Farkas L, Farkas D, Gauldie J et al. Transient overexpression of Gremlin results in epithelial activation and reversible fibrosis in rat lungs. Am J Respir Cell Mol Biol 2011;44:870–878.
Myllarniemi M, Lindholm P, Ryynanen MJ et al. Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis. Am J Respir Crit Care Med 2008;177:321–329.
Botelho FM, Rangel-Moreno J, Fritz D et al. Pulmonary expression of oncostatin M (OSM) promotes inducible BALT formation independently of IL-6, despite a role for IL-6 in OSM-driven pulmonary inflammation. J Immunol 2013;191:1453–1464.
Fritz DK, Kerr C, Tong L et al. Oncostatin-M up-regulates VCAM-1 and synergizes with IL-4 in eotaxin expression: involvement of STAT6. J Immunol 2006;176:4352–4360.
Schust J, Sperl B, Hollis A et al. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006;13:1235–1242.
Volk SW, Wang Y, Mauldin EA et al. Diminished type III collagen promotes myofibroblast differentiation and increases scar deposition in cutaneous wound healing. Cells Tissues Organs 2011;194:25–37.
Merkel JR, DiPaolo BR, Hallock GG et al. Type I and type III collagen content of healing wounds in fetal and adult rats. Proc Soc Exp Biol Med 1988;187:493–497.
Ray S, Ju X, Sun H et al. The IL-6 trans-signaling-STAT3 pathway mediates ECM and cellular proliferation in fibroblasts from hypertrophic scar. J Invest Dermatol 2013;133:1212–1220.
Pechkovsky DV, Prêle CM, Wong J et al. STAT3-mediated signaling dysregulates lung fibroblast-myofibroblast activation and differentiation in UIP/IPF. Am J Pathol 2012;180:1398–1412.
Kida H, Mucenski ML, Thitoff AR et al. GP130-STAT3 regulates epithelial cell migration and is required for repair of the bronchiolar epithelium. Am J Pathol 2008;172:1542–1554.
Masterson JC, Molloy EL, Gilbert JL et al. Bone morphogenetic protein signalling in airway epithelial cells during regeneration. Cell Signal 2011;23:398–406.
Pegorier S, Campbell GA, Kay AB et al. Bone morphogenetic protein (BMP)-4 and BMP-7 regulate differentially transforming growth factor (TGF)-beta1 in normal human lung fibroblasts (NHLF). Respir Res 2010;11:85.
Izumi N, Mizuguchi S, Inagaki Y et al. BMP-7 opposes TGF-beta1-mediated collagen induction in mouse pulmonary myofibroblasts through Id2. Am J Physiol Lung Cell Mol Physiol 2006;290:L120–L126.
Pope SM, Zimmermann N, Stringer KF et al. The eotaxin chemokines and CCR3 are fundamental regulators of allergen-induced pulmonary eosinophilia. J Immunol 2005;175:5341–5350.
Pope SM, Fulkerson PC, Blanchard C et al. Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J Biol Chem 2005;280:13952–13961.
Viola JP, Kiani A, Bozza PT et al. Regulation of allergic inflammation and eosinophil recruitment in mice lacking the transcription factor NFAT1: role of interleukin-4 (IL-4) and IL-5. Blood 1998;91:2223–2230.
Atochina EN, Beers MF, Tomer Y et al. Attenuated allergic airway hyperresponsiveness in C57BL/6 mice is associated with enhanced surfactant protein (SP)-D production following allergic sensitization. Respir Res 2003;4:15.
Bringardner BD, Baran CP, Eubank TD et al. The role of inflammation in the pathogenesis of idiopathic pulmonary fibrosis. Antioxid Redox Signal 2008;10:287–301.
Abdelaziz MM, Samman YS, Wali SO et al. Treatment of idiopathic pulmonary fibrosis: is there anything new? Respirology 2005;10:284–289.
Camelo A, Dunmore R, Sleeman MA et al. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol 2014;4:173.
Nathan N, Thouvenin G, Fauroux B et al. Interstitial lung disease: physiopathology in the context of lung growth. Paediatr Respir Rev 2011;12:216–222.
Nightingale J, Patel S, Suzuki N et al. Oncostatin M, a cytokine released by activated mononuclear cells, induces epithelial cell-myofibroblast transdifferentiation via Jak/Stat pathway activation. J Am Soc Nephrol 2004;15:21–32.
Pollack V, Sarközi R, Banki Z et al. Oncostatin M-induced effects on EMT in human proximal tubular cells: differential role of ERK signaling. Am J Physiol Renal Physiol 2007;293:F1714–F1726.
Acknowledgements
We thank Christine Demers (McMaster University) for excellent technical assistance and Sean Lauber (McMaster University) for his expertise. This study was supported by operating Grant 102562 from the Canadian Institute of Health Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Wong, S., Botelho, F., Rodrigues, R. et al. Oncostatin M overexpression induces matrix deposition, STAT3 activation, and SMAD1 Dysregulation in lungs of fibrosis-resistant BALB/c mice. Lab Invest 94, 1003–1016 (2014). https://doi.org/10.1038/labinvest.2014.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2014.81
This article is cited by
-
Role of oncostatin M in the pathogenesis of vernal keratoconjunctivitis: focus on tissue remodeling
Japanese Journal of Ophthalmology (2021)
-
The role of the oncostatin M/OSM receptor β axis in activating dermal microvascular endothelial cells in systemic sclerosis
Arthritis Research & Therapy (2020)
-
Oncostatin M exerts a protective effect against excessive scarring by counteracting the inductive effect of TGFβ1 on fibrosis markers
Scientific Reports (2019)
-
Overexpression of OSM and IL-6 impacts the polarization of pro-fibrotic macrophages and the development of bleomycin-induced lung fibrosis
Scientific Reports (2017)
-
Bidirectional role of IL-6 signal in pathogenesis of lung fibrosis
Respiratory Research (2015)