Abstract
Transforming growth factor-β1 (TGF-β1) is the master cytokine in the pathogenesis of liver fibrosis. TGF-β1 and extent of fibrosis were correlated recently to the serpin SERPINB3 in idiopathic pulmonary fibrosis, a chronic disease recalling liver cirrhosis. The aim of this study was to assess the relation between SERPINB3, TGF-β1 and fibrosis in chronic liver diseases and to determine the effect of this serpin on TGF-β1 expression using in vitro models. SERPINB3 and TGF-β1 were evaluated in liver biopsies of 94 patients with chronic liver disease. The effect of SERPINB3 on TGF-β1 expression was determined in primary human hepatocytes, HepG2 and Huh7 cells transfected with intact SERPINB3 human gene or with reactive site loop deleted mutants. A significant correlation between TGF-β1 and SERPINB3 at the protein level was observed in liver biopsies, confirmed by a positive correlation at mRNA level. Both proteins were correlated to the extent of liver fibrosis. All transfected cells showed increased TGF-β1 mRNA and protein production and the integrity of the reactive site loop of the serpin was crucial to achieve this effect. In conclusion, chronically damaged hepatocytes produce SERPINB3 and TGF-β, and the anti-protease activity of this serpin might be implicated in TGF-β1 induction.
Similar content being viewed by others
Main
Hepatic fibrosis is the result of the wound-healing response of the liver to repeated injury, resulting from chronic damage. Repair mechanisms lead to replacement of injured parenchymal cells by connective tissue with progressive accumulation of extracellular matrix (ECM) proteins, including fibrillar collagen type I and III, which is characteristic of most types of chronic liver disease.1, 2 If the injury persists, the continuous fibrotic process leads to the formation of bands of collagen, bridging fibrosis and frank cirrhosis with liver failure, and portal hypertension. In addition, epidemiological data have shown that liver cirrhosis, regardless of its etiology, is the most important risk factor for the development of primary liver cancer.3, 4
The fibrogenic mechanism is the result of the interplay of several pro- and anti-fibrotic/inflammatory cytokines.5 The pro-fibrogenic growth factors include platelet-derived growth factor and transforming growth factor-β (TGF-β), which is the fibrogenic master cytokine.6 TGF-β represents a large family of growth and differentiation factors that mobilize complex signaling networks to regulate cellular differentiation, proliferation, motility, adhesion, and apoptosis. TGF-β acts via transmembrane receptors, eg, the type I, TGF-β receptor, to intracellular mediators of the Smad family that are phosphorylated on serine residues and form heteromeric complexes with a common mediator, Smad4. This complex translocates to the nucleus, where Smads regulate gene expression either directly or in association with a number of coactivators and corepressors. TGF-β also can be involved in other pathways, eg, the mitogen-activated protein kinase pathways,7 although the mechanisms of activation seem to be different. Cross-talk between the Smad signaling cascade and other pathways also adds complexity to the system.8, 9
In mammals, three isoforms termed TGF-β 1, 2, and 3 have been identified and are synthesized as large latent precursors that are unable to trigger signaling via high-affinity TGF-β receptors unless cleavage of the active C-terminal dimer occurs, with extracellular dissociation.10 Among the three isoforms, the subtype TGF-β1 has been implicated particularly in the pathogenesis of liver fibrosis by inducing production of ECM proteins, while inhibiting the synthesis of matrix-degrading proteolytic enzymes.10 Inflammatory and mesenchymal cells, including hepatic stellate cells, which are also one of the most important targets of activated TGF-β1, have been identified as the principal source of this cytokine.6, 11 However, growing evidence also supports the ability of parenchymal hepatocytes to produce TGF-β1, both in chronic liver disease12, 13, 14 and in cultured hepatocytes.15, 16
Recently, Calabrese et al17 reported a strict correlation between TGF-β and squamous cell carcinoma antigen (SCCA) expression in the parenchymal lung tissue of patients with idiopathic pulmonary fibrosis. The key pathological features of this disease resemble the pathogenesis of cirrhosis, including epithelial damage, abnormal mesenchymal cell activation, and proliferation with excess of ECM deposition, driven by TGF-β1 as pivotal cytokine.18 SERPINB3 (SCCA1) and SERPINB4 (SCCA2), the two isoforms of SCCA, are members of the ovalbumin family of serine proteinase inhibitors.19 Serpins function via an exposed reactive site loop (RSL) of about 20 amino acids and inhibit irreversibly proteinases through a suicide substrate inhibition mechanism.19
SCCA isoforms have been found overexpressed in epithelial tumors,20 primary liver cancer,21, 22, 23 and chronically damaged hepatocytes.24, 25, 26
The aim of this study was to assess the potential correlation between TGF-β1 and SERPINB3 in chronically damaged human liver tissue and to determine their relationship using in vitro models of cultured cells transfected with vectors expressing SERPINB3.
MATERIALS AND METHODS
Patients
The study was carried out in 94 consecutive patients with chronic liver disease (63 with chronic HCV infection, 17 with chronic anti-HBe positive HBV infection, and 14 patients with alcoholic liver disease) in whom percutaneous liver biopsies were performed for diagnostic purposes, under informed consent. Histologic features revealed the presence of chronic hepatitis in 65 patients and of liver cirrhosis in 29 patients. Epidemiologic and clinical characteristics of the patients included in the study are reported in Table 1. Formalin-fixed and paraffin-embedded sections were available in all cases. Control liver biopsies were obtained from 20 patients who underwent cholecystectomy or liver biopsy for the staging of lymphoproliferative disease.14
Histological Evaluation
Sections (4–8 μm) from formalin-fixed and paraffin-embedded liver biopsies were stained with hematoxylin-eosin. Histological grading and staging was assessed using the Ishak scoring system27 by two pathologists. For immunohistochemical analyses, two consecutive liver sections of all histological samples were subjected to antigen retrieval by heating in a microwave oven on high power for 8 min in 0.01 mol/l citrate buffer (pH 6.0), followed by incubation with a mouse monoclonal anti-TGF-β1 primary antibody (150 μg/ml; dilution 1:20, Genzyme Diagnostics, Cambridge, MA, USA) or with a polyclonal anti-SCCA antibody (4μg/ml) (Xeptagen S.p.A, Marghera, VE, Italy), recognizing both SERPINB3 and SERPINB4 isoforms.
Before incubation with primary antibody, sections were treated with a biotin blocking kit (Vector Peterborough, UK) to inhibit endogenous biotin. Immune staining was then carried out using the Vectastain ABC kit (Vector Peterborough) with 3,3-diaminobenzidine tetrahydrochloride as the chromogenic substrate. Sections were counterstained with Mayer's hematoxylin. Negative controls were performed for all immunohistochemistry experiments, including incubation with the omission of primary or secondary antibody, or with normal serum to control for background reactivity. The extent of the immunoreactivity in parenchymal liver cells, defined as the mean percentile value of immunoreactive cells, independently obtained by two pathologists, was semiquantitatively scored as follows: 0=absence of immunoreactive cells; 1=1–10% immunoreactive cells; 2=11–50% immunoreactive cells; 3=>50% immunoreactive cells. Intra- and inter-observer differences were <5% and discordant cases were re-evaluated by the two observers.
Construction of SERPINB3 Expression Vectors
To obtain SERPINB3 RNA for plasmid construction, total RNA was extracted from human liver cancer tissue using the Trizol reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer's instructions. The first strand of cDNA was synthesized (Superscript II reverse transcriptase, Invitrogen) and used for human wild-type SERPINB3 (GenBank NM_006919) cDNA gene amplification by nested polymerase chain reaction (PCR). The PCR fragment was cloned directly into the mammalian expression vector pcDNA3.1D/V5-His-TOPO (Invitrogen). The construct was propagated into Escherichia coli TOP10 competent cells (Invitrogen) and purified using the Genopure Plasmid Maxi kit (Roche Applied Science, Indianapolis, USA). pcDNA3.1D/V5-His/lacZ vector, lacking the SERPINB3 insert, was used as a negative control. Sequence controls were carried out using an ABI 310 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). To obtain SERPINB3/GFP vector for immunofluorescence analysis, the PCR fragment of SERPINB3 was cloned into the mammalian expression vector pcDNA3.1/NT-GFP-TOPO (Invitrogen) and propagated and purified as mentioned earlier.
To explore the effect of the RSL of SERPINB3 in the induction of the TGF-β1 gene, previously described deletion loop mutants of SERPINB3,28 were also used for cell transfection. The following vectors showing lack of inhibition of target proteinases were chosen: Mutant A (deletion of total hinge region of RSL-SERPINB3), Mutant B (deletion of total RSL-SERPINB3), and Mutant C (Mutation of the P14 codon from alanine to arginine) (Figure 6).
Hepatic Cells Culture and Reagents
Human hepatoblastoma (HepG2) and Huh7 cell lines (kindly provided by Professor G Giannelli, University of Bari, Italy) were cultured in minimum essential medium (MEM; Sigma-Aldrich, St Louis, MO, USA) and in Dulbecco's Modified Medium (Sigma-Aldrich), respectively, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 200 mM L-glutamine and 1 × MEM non-essential amino acid solution (Sigma-Aldrich) at 37°C in a humidified atmosphere containing 5% CO2.
Cryopreserved primary human hepatocytes (HHEP) were purchased by Provitro (GmbH, Berlin, Germany) and were cultured using the recommended standard culture medium (Provitro GmbH, Germany) at 37°C in a humidified atmosphere containing 5% CO2.
Hepatic Cells Transfection
HepG2, Huh7 cell lines and primary hepatocytes were transiently transfected with plasmid vectors carrying the SERPINB3 gene, or SERPINB3 deletion mutants, whereas cells transfected with the empty vector were used as a control. Cells were plated in six-well plates at a density of 1 × 106 and of 8 × 105 cells/well (∼60% confluence), respectively, 24 h before transfection. Cells were then incubated with 1 μg of plasmid DNA/well in the presence of Lipofectamin Reagent and Plus Reagent (Invitrogen). After 5 h incubation at 37°C in a humidified atmosphere containing 5% CO2, wells were washed twice with OptiMEM (Invitrogen) and incubated with fresh RPMI medium (Sigma-Aldrich) supplemented with 10% FBS for a further 48 h.
SERPINB3 and TGF-β1 mRNA Expression
Total RNA was extracted using RNasy Trizol (Invitrogen) according to the manufacturer's instructions and quantified by spectrophotometry at 260 nm. Total RNA (up to 1 μg) was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) in a reaction mix consisting of: 4 μl of 5X buffer, 2 μl DTT (0.1 M), 2 μl dNTPs (5 mM), 1 μl of primers oligo dT (500 μg/ml), 1 μl (200 U) Superscript II and 1 μl (40 U) RNase inhibitor (Invitrogen).
Expression of TGF-β1 was determined using SYBR green master mix (Roche Diagnostics GmbH, Indianapolis, USA) and the following primers: TGF-β1 sense: 5′-AAGTGGACATCAACGGGTTC-3′; TGFβ1 reverse 5′-GTCCTTGCGGAAGTCAATGT-3′; for SERPINB3 mRNA quantification: sense primer 5′-GCA AAT GCT CCA GAA GAA AG-3′, reverse primer 5′-CGA GGC AAA ATGAAAA AGA TG-3′.
The housekeeping gene Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (sense: 5′-TGG TAT TCG GGA AGG ACT CAT GAC-3′, reverse: 5′-ATG CCA GTG AGC TTC CCG TTC AGC-3′) was determined in parallel in all the amplification sets to assess the integrity of total RNA extracts.
The single-tube RT-PCR assays were performed using the LightCycler instrument (Roche Diagnostics GmbH) and consisted of 1 denaturation cycle at 95°C for 10 min, 45 cycles of amplification at 94°C for 1 s, 62°C for 10 s, and 72°C for 10 s. Melt curve analysis was performed by ramping products from 45 to 98°C, acquiring fluorescence readings for each degree change. For SERPINB3 and TGF-β1 gene, the fluorescence of the SYBR green dye was determined as a function of the PCR cycle number, giving the threshold cycle (CT) number. The CT values were used to quantify the PCR product, ΔCT was calculated by subtracting CT (control gene: GAPDH) from CT (target gene: TGF-β1). The ΔCT value of the control (cells transfected with empty vector) was used arbitrarily as a constant that was subtracted from all other ΔCT values to determine ΔΔCT value. Samples were run in triplicate, and fold changes were generated for each sample by calculating .29
Specificity of the amplified PCR products was determined by melting curve analysis and confirmed by agarose gel electrophoresis and ethidium bromide staining.
Protein Expression
Immunofluorescence
The expression of SERPINB3 and TGF-β1 in transiently transfected cells was assessed by immunofluorescence. HepG2 and primary human hepatocytes were seeded on slides (2 × 105 cells cells/slide), transfected with SERPINB3-GFP, and cultured for 48 h.
Cells were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X100 and blocked with 5% BSA in PBS. Slides were then incubated at room temperature for 2 h with a mouse monoclonal anti-TGF-β antibody (NovoCastra, Newcastle, UK) as described earlier,21, 30 washed with 0.1% Tween 20 in PBS and incubated with the TRITC-conjugated secondary antibody (1:50 dilution) (Dako, Copenhagen, Denmark) at room temperature for 2 h. The cellular nuclei were stained by incubation with 20 μg/ml of Hoechst 33342 (Sigma-Aldrich) for 5 min. Slides were washed in PBS, mounted with glycerol (Sigma-Aldrich), and the results were observed on fluorescence microscopy (Axiovert 200 M, Carl Zeiss MicroImaging GmbH, Göttingen, Germany).
ELISA assay
The presence of biologically active TGF-β1 in the supernatant of transfected cells was determined using the TGF-β1 Emax ImmunoAssay system, following the manufacturer's instructions (Promega Corporation, Madison, WI, USA).
Biological activity was quantified using standard curves with a linear range between 15.6 and 1000 pg/ml.
Statistical Analysis
Statistical significance was determined by non-parametric procedures using the unpaired t test, Welch corrected, Mann–Whitney test and Fisher's exact test. Normality of distribution for quantitative variables was assessed by Kolmogorov and Smirnov test. To evaluate simple linear relationships between quantitative variables, Spearman's correlation coefficients were applied, when indicated. All tests were two sided. The calculations were carried out with Graph Pad InStat Software (San Diego, CA, USA). The null hypothesis was rejected at P<0.05.
RESULTS
Immunohistochemical Findings
In liver specimens of patients with chronic hepatitis, parenchymal TGF-β1 was detected usually in the cytoplasm of hepatocytes at the interface of the portal space and in close proximity to areas of fibrosis, whereas in cases with cirrhosis, a clusters of positive cells were observed both in hepatocytes at the border between the parenchyma and connective tissue and entrapped in fibrotic bands surrounding the pseudolobules.
Immunostaining with an anti-SCCA antibody, performed in sequential liver sections, showed a distribution pattern similar to that of TGF-β1 (Figure 1). Although SCCA immunoreactivity detected by the polyclonal anti-SCCA antibody could not exclude also the presence of the SERPINB4 isoform, SERPINB3 overexpression was confirmed at mRNA level, using specific primers, in 9/12 available frozen liver biopsies from patients with chronic hepatitis (Supplementary Figure 1).
SCCA and TGF-β1 co-localization in the liver. Example of immunostaining for SCCA isoforms, including SERPINB3 and SERPINB4 (a) and for TGF-β (b) performed in sequential sections of a liver biopsy of a cirrhotic patient. Immune reactivity showed a similar pattern of distribution, characterized by clusters of positive cells. Magnification 30 × .
The correlation of the two molecules was also supported by the distribution of TGF-β1 scores in liver biopsies divided on the basis of SCCA score (<2 vs ≥2), and the results were independent of the etiology of chronic liver disease (Figure 2a). Moreover, a similar behavior was observed in HCV patients infected with different genotypes (Figure 2b). Neither TGF-β1 nor SCCA immunoreactivity were detectable in normal liver tissue samples used as controls.
Relationship between SCCA and TGF-β1 expression in the liver. (a) Immunoreactivity in relation to liver disease etiology. Distribution of TGF-β1 mean score values, determined in liver biopsies of the study population grouped by etiology and divided on the basis of the extent of SCCA score (<2 vs ≥2) evaluated in sequential sections of the same liver specimen. (b) Immunoreactivity in relation to HCV genotype. Distribution of mean score values of TGF-β1 in liver biopsies of all HCV-infected patients grouped by HCV genotype (genotype 1–3) and divided on the basis of the extent of the corresponding SCCA score (<2 vs ≥2). Statistical analyses were performed using the Student's t test Welch's corrected. **SCCA score <2 vs ≥2=(P<0.005); *** SCCA score <2 vs ≥2=(P<0.001).
Correlation with Liver Fibrosis
To define better the correlation of the serpin and of TGF-β1 with the extent of liver fibrosis, liver biopsies of patients with chronic hepatitis were analyzed for both molecules in relation to the corresponding fibrosis score. As shown in Figure 3, Spearman analysis confirmed the positive correlation of SCCA (Figure 3a) and of TGF-β (Figure 3b) scores with the fibrosis score (r=0.6485, P<0.0001 and r=0.7424, P<0.0001, respectively). Clinical and biochemical parameters of the study population, including age, sex, and transaminases, did not show any correlation with the extent of expression of TGF-β1 or SCCA proteins.
Correlation of fibrosis score with SCCA (a) and TGF-β1 (b). The parameters were evaluated in individual patients with chronic hepatitis. Fibrosis staging27 was assessed in formalin-fixed and paraffin-embedded liver biopsy sections stained with hematoxylin-eosin. Statistical analysis was performed using Spearman correlation. A positive linear correlation between the degree of expression of both SCCA (r=0.6485, 95% confidence interval=0.4716–0.7752, P<0.0001) and TGF-β (r=0.7424, 95% confidence interval=0.6014–0.8386, P<0.0001) and the extent of fibrosis score was observed.
In Vitro Studies
TGF-β in cells transfected with SERPINB3
To investigate whether SERPINB3 could affect TGF-β1 expression, we used an in vitro model in which different cell lines and primary human hepatocytes were transfected with the human SERPINB3 gene.
Total RNA was purified from transiently transfected HepG2, Huh 7 cells and human hepatocytes and from the corresponding control cells transfected with the pcDNA3.1 empty vector. SERPINB3 mRNA quantification was then assessed in each sample by real-time RT-PCR using primers specific for the human SERPINB3 gene. TGF-β1 mRNA expression showed a significant increase of this cytokine in all SERPINB3-transfected cells at 48 h from transfection, but not in control cells (Figure 4a).
TGF-β1 induction in cells transiently transfected with SERPINB3. (a) TGF-β1 mRNA induction in HepG2, Huh7 cells, and primary human hepatocytes. TGF-β1 transcripts were assessed in each sample of transiently transfected cells at 48 h from the transfection with SERPINB3 (gray bars) or with the empty vector (white bars) by real-time PCR using primers specific for the human TGF-β1 gene. Changes in mRNA gene expression were reported as fold changes relative to controls (untransfected cells at the corresponding time) using the method.29 The results were normalized to the GAPDH housekeeping gene. Results are expressed as mean±s.e.m. of three independent experiments. Statistical significance was determined using the Student's t test Welch's corrected. *P<0.05. (b) TGF-β1 protein concentrations in supernatant of transfected cells. Distribution of biologically active TGF-β1 protein concentrations in the supernatant of the corresponding cell lines and primary human hepatocytes. Results are representative of three independent experiments and are expressed as mean±s.e.m. Statistical analysis was performed using Unpaired t test with Welch's correction **P<0.005; ***P<0.001.
To confirm the induction of TGF-β1 by SERPINB3 not only at the transcription level, but also at protein level, biologically active TGF-β1 was determined in parallel by ELISA in the supernatant of transiently transfected cells using the Emax ImmunoAssay Elisa test. As reported in Figure 4b, a similar increase of the cytokine production at the corresponding time was observed.
Localization of SERPINB3 and TGF-β1 by immunofluorescence
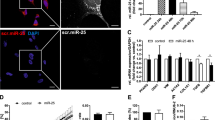
To characterize the cellular localization of TGF-β1 and SERPINB3, HepG2 cells and primary human hepatocytes (HHEP) transiently transfected with SERPINB3-GFP were analyzed by immunofluorescence, where cell nuclei were counterstained with Hoechst.
As shown in Figure 5, transfected HepG2 and HHEP cells exhibited high levels of TGF-β1, co-localizing with SERPINB3, whereas weak TGF-β1 and SERPINB3 reactivity was detectable in untransfected cells.
Immunofluorescence analysis in SERPINB3-GFP-transfected cells. SERPINB3 (green) and TGF-β1 localization (red) were detected by immunofluorescence. Cell nuclei were counterstained with Hoechst 33342 (blue) and shown as merge with the SERPINB3 and TGF-β1 immunofluorescences. A weak TGF-β1 and SERPINB3 reactivity was detectable in untransfected HepG2 cells and in primary human hepatocytes (HHEP), whereas a remarkable TGF-β1 fluorescence signal was visible in SERPINB3-transfected cells, with a similar pattern of distribution in both cell types. Original magnification, 40 × .
The fluorescence signals and the pattern of expression of TGF-β1 were comparable in HepG2 cells and in human hepatocytes expressing SERPINB3.
Influence of the SERPINB3 RSL in TGF-β1 gene expression
To evaluate the requirement for the RSL of SERPINB3 in affecting TGF-β1 expression, cell transfection was carried out with a panel of RSL mutants. The amount of TGF-β1 mRNA in HepG2 cells after 48 h of transfection was strongly dependent on the integrity of the reactive loop. As shown in Figure 6, a lack of TGF-β1 mRNA induction was observed with Mutant A and Mutant B, containing large deletions of the RSL of SERPINB3, whereas reduced TGF-β1 mRNA induction was achieved with Mutant C, which contains a single amino-acid substitution within the RSL.
TGF-β1 mRNA in HepG2 cells transfected with SERPINB3 reactive site loop mutants. Upper panel: TGF-β mRNA levels after 48 h transfection with plasmids containing the entire SERPINB3 sequence or RLS mutants. Transfection with RSL mutants determined abrogation of TGF-β1 mRNA induction and the maximal effect was observed after transfection with plasmid constructs carrying the larger reactive site loop deletions (mutants A and B), while low TGF-β1 mRNA induction was detected with mutant C, which contains a single amino-acid substitution in the reactive site loop. Lower panel: SERPINB3 partial sequence. The RSL from P15-P5′ (Schechter and Berger numbering)42 is underlined. Mutant A: deletion of total hinge region of RSL; mutant B: deletion of total RSL; mutant C: AlaP14Arg.
DISCUSSION
Liver fibrogenesis is the common response of the liver to toxic, infectious, or metabolic injuries and is characterized by excessive accumulation of ECM caused by both increased synthesis and deposition of newly formed components and decreased or unbalanced degradation of ECM elements.31 In the evolution of cirrhosis, an imbalanced immunomodulation occurs, characterized by the interplay of many pro- and anti-fibrotic/-inflammatory cytokines.32 Knowledge of the fine molecular mechanisms underlying this pathological process is of great clinical importance, because it might influence targeted therapeutic approaches. A central role in liver fibrogenesis has been assigned to TGF-β1, widely recognized as the pivotal pro-fibrogenic agent in chronic liver injury.6 Non-parenchymal liver cells are considered the main source of this cytokine, however, several clinical and experimental findings support the ability of hepatocytes to produce TGF-β1 in pathological conditions.14, 15 TGF-β1 not only enhances ECM synthesis but also inhibits ECM degradation by downregulating matrix-degrading enzymes and promoting expression of matrix metalloproteinase (MMP) inhibitors.33, 34 Proteolytic mechanisms are indeed additional players involved in matrix remodeling and crucial roles of cysteine and serine proteases and their inhibitors have been recognized not only for tissue remodeling, but also for angiogenesis, stromal invasion, and formation of metastases by malignant cancer cells.35, 36
In this study, we have confirmed that TGF-β1 is expressed in chronically damaged hepatocytes, and we have shown for the first time that the same cells can synthesize SCCA isoforms, whereas both molecules are not expressed in normal human liver. Although SERPINB4 isoform could not be excluded, the presence of SERPINB3 overexpression in liver biopsies was confirmed at mRNA level. The positive correlation found between the extent of expression of both TGF-β1 and SERPINB3 and fibrosis grade suggests that both molecules are involved in this process, leading to progressive collagen deposition, as a defence mechanism following hepatic injury. This ov-serpin has been shown to render cells more resistant to apoptotic cell death,37, 38 and the pro-apoptotic effect of TGF-β1 in hepatocytes39 might represent a counterbalancing mechanism, leading to accumulation of ECM as a secondary effect. More recently, epithelial mesenchymal transition capacity, typically described for TGF-β1,40 has been also ascribed to SERPINB3.41 These cellular features acquired by damaged hepatocytes can explain, at least in part, the higher risk of neoplastic transformation found in liver cirrhosis, when this pathological condition can be considered the end stage of a chronic inflammatory process. Both SERPINB3 and SERPINB4 have been indeed found overexpressed in hepatocellular carcinoma21, 22 and in preneoplastic liver lesions.26
Significant increase of TGF-β1 in SERPINB3-transfected cell lines and primary human hepatocytes was documented both at mRNA level and by its increased protein production. The integrity of the RSL was crucial to determine this effect, as shown by the results obtained in cells transfected with RSL-deleted mutants. The abrogation of TGF-β1 increase in cells transfected with plasmid vectors containing large RSL deletions suggest that the anti-protease activity of the serpin was implicated in TGF-β1 induction. These findings are in keeping with recent results, obtained in the A549 lung epithelial cell line, where the addition of exogenous recombinant SERPINB3 protein induced an increase of TGF-β1 mRNA expression and further support the role of SERPINB3 in fibrogenesis.17 The parallel expression of the two molecules in idiopathic pulmonary fibrosis and in chronically damaged livers provides evidence that a common fibrogenic mechanism is likely to occur, where SERPINB3 might act as a TGF-β upstream modulator protein.
In conclusion, this study has provided evidence that chronically damaged hepatocytes produce SERPINB3 and TGF-β1 and that the anti-protease activity of this serpin could be implicated in TGF-β1 induction.
Accession codes
References
Friedman SL . Liver fibrosis from bench to bedside. J Hepatol 2003;38:38–53.
Bataller R, Brenner DA . Liver fibrosis. J Clin Invest 2005;115:209–218.
Bruix J, Llovet JM . Hepatitis B virus and hepatocellular carcinoma. J Hepatol 2003;39:S59–S63.
Benvegnù L, Gios M, Boccato S, et al. Natural history of compensated viral cirrhosis: a prospective study on the incidence and hierarchy of major complications. Gut 2004;53:744–749.
Pinzani M, Rombouts K . Liver fibrosis: from the bench to clinical targets. Dig Liver Dis 2004;36:231–242.
Bissell DM, Roulot D, George J . Transforming growth factor beta and the liver. Hepatology 2001;34:859–867.
Moustakas A, Heldin CH . Non-Smad TGF-beta signals. J Cell Sci 2005;118:3573–3584.
Zhang Y, Feng XH, Derynck R . Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 1998;394:909–913.
Liberati NT, Datto MB, Frederick JP, et al. Smads bind directly to the Jun family of AP-1 transcription factors. Proc Natl Acad Sci USA 1999;96:4844–4849.
Miyazono K, Hellman U, Wernstedt C, et al. Latent high molecular weight complex of transforming growth factor beta 1. Purification from human platelets and structural characterization. J Biol Chem 1988;263:6407–6415.
Knittel T, Janneck T, Müller L, et al. Transforming growth factor beta 1-regulated gene expression of Ito cells. Hepatology 1996;24:352–360.
Nelson DR, Gonzalez-Peralta RP, Qian K, et al. Transforming growth factor-beta 1 in chronic hepatitis C. J Viral Hepat 1997;4:29–35.
Dudás J, Kovalszky I, Gallai M, et al. Expression of decorin, transforming growth factor-beta 1, tissue inhibitor metalloproteinase 1 and 2, and type IV collagenases in chronic hepatitis. Am J Clin Pathol 2001;115:725–735.
Calabrese F, Valente M, Giacometti C, et al. Parenchymal transforming growth factor beta-1: its type II receptor and Smad signaling pathway correlate with inflammation and fibrosis in chronic liver disease of viral etiology. J Gastroenterol Hepatol 2003;18:1302–1308.
Nitta T, Kim JS, Mohuczy D, et al. Murine cirrhosis induces hepatocyte epithelial mesenchymal transition and alterations in survival signaling pathways. Hepatology 2008;48:909–919.
Dooley S, Hamzavi J, Ciuclan L, et al. Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008;135:642–659.
Calabrese F, Lunardi F, Giacometti C, et al. Overexpression of squamous cell carcinoma antigen in idiopathic pulmonary fibrosis: clinicopathological correlations. Thorax 2008;63:795–802.
Selman M, King TE, Pardo A . Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 2001;134:136–151. Review.
Gettins PG . Serpin structure, mechanism, and function. Chem Rev 2002;102:4751–4804. Review.
Kato H . Expression and function of squamous cell carcinoma antigen. Anticancer Res 1996;16:2149–2153.
Pontisso P, Calabrese F, Benvegnù L, et al. Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Br J Cancer 2004;90:833–837.
Giannelli G, Marinosci F, Sgarra C, et al. Clinical role of tissue and serum levels of SCCA antigen in hepatocellular carcinoma. Int J Cancer 2005;116:579–583.
Giannelli G, Fransvea E, Trerotoli P, et al. Clinical validation of combined serological biomarkers for improved hepatocellular carcinoma diagnosis in 961 patients. Clin Chim Acta 2007;383:147–152.
Beneduce L, Castaldi F, Marino M, et al. Squamous cell carcinoma antigen-immunoglobulin M complexes as novel biomarkers for hepatocellular carcinoma. Cancer 2005;103:2558–2565.
Biasiolo A, Chemello L, Quarta S, et al. Monitoring SCCA-IgM complexes in serum predicts liver disease progression in patients with chronic hepatitis. J Viral Hepat 2008;15:246–249.
Guido M, Roskams T, Pontisso P, et al. Squamous cell carcinoma antigen in human liver carcinogenesis. J Clin Pathol 2008;61: 445–447.
Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol 1995;22:696–699.
Moore PL, Ong S, Harrison TJ . Squamous cell carcinoma antigen 1-mediated binding of hepatitis B virus to hepatocytes does not involve the hepatic serpin clearance system. J Biol Chem 2003;278:46709–46717.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402–408.
Beghe B, Bazzan E, Baraldo S, et al. Transforming growth factor-beta type II receptor in pulmonary arteries of patients with very severe COPD. Eur Respir J 2006;28:556–562.
Friedman SL . Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem 2000;275:2247–2250. Review.
Gressner AM . Mediators of hepatic fibrogenesis. Hepato-Gastroenterol 1996;43:92–103.
Ogawa K, Chen F, Kuang C, et al. Suppression of matrix metalloproteinase-9 transcription by transforming growth factor-beta is mediated by a nuclear factor-kappaB site. Biochem J 2004;381: 413–422.
Verrecchia F, Chu ML, Mauviel A . Identification of novel TGF-beta /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem 2001;276:17058–17062.
Liotta LA, Stetler-Stevenson WG . Tumor invasion and metastasis: an imbalance of positive and negative regulation. Cancer Res 1991;51:5054–5059. Review.
He Y, Liu XD, Chen ZY, et al. Interaction between cancer cells and stromal fibroblasts is required for activation of the uPAR-uPA- MMP-2 cascade in pancreatic cancer metastasis. Clin Cancer Res 2007;13:3115–3124.
Suminami Y, Nagashima S, Vujanovic NL, et al. Inhibition of apoptosis in human tumor cells by tumor-associated serpin, SCC antigen. Br J Cancer 2000;82:981–989.
Takeda A, Kajiya A, Iwasawa A, et al. Aberrant expression of serpin squamous cell carcinoma antigen 2 in human tumor tissues and cell lines: evidence of protection from tumor necrosis factor-mediated apoptosis. Biol Chem 2002;383:1231–1236.
Nguyen LN, Furuya MH, Wolfraim LA, et al. Transforming growth factor-beta differentially regulates oval cell and hepatocyte proliferation. Hepatology 2007;45:31–41.
Xu J, Lamouilee S, Derynck R . TGF-β- induced epithelial to mesenchymal transition. Cell Res 2009;19:156–172.
Quarta S, Turato C, Ruvoletto MG, et al. SERPINB3 induces epithelial mesenchymal transition. Hepatology 2009;50 (Suppl):379A.
Schechter I, Berger A . On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 1967;27:157–162.
Acknowledgements
This work was supported in part by a grant from the National Ministry of Education, University and Research (FIRB Project Prot. RBLA03S4SP_005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Supplementary information
Rights and permissions
About this article
Cite this article
Turato, C., Calabrese, F., Biasiolo, A. et al. SERPINB3 modulates TGF-β expression in chronic liver disease. Lab Invest 90, 1016–1023 (2010). https://doi.org/10.1038/labinvest.2010.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2010.55
Keywords
This article is cited by
-
Enhanced Wnt Signalling in Hepatocytes is Associated with Schistosoma japonicum Infection and Contributes to Liver Fibrosis
Scientific Reports (2017)
-
SerpinB3 Promotes Pro-fibrogenic Responses in Activated Hepatic Stellate Cells
Scientific Reports (2017)
-
Hepatic progenitor cells express SerpinB3
BMC Cell Biology (2014)
-
SERPINB3 is associated with TGF-β1 and cytoplasmic β-catenin expression in hepatocellular carcinomas with poor prognosis
British Journal of Cancer (2014)
-
SERPINB3 is associated with longer survival in transgenic mice
Scientific Reports (2013)