Abstract
To maintain normal retinal function, retinal pigment epithelial (RPE) cells engulf photoreceptor outer segments (ROS) enriched in free fatty acids (FFAs). We have previously demonstrated fatty acid-binding protein 5 (FABP5) downregulation in the RPE/choroidal complex in a mouse model of aging and early age-related macular degeneration. FABPs are involved in intracellular transport of FFAs and their targeting to specific metabolic pathways. To elucidate the role of FABP5 in lipid metabolism, the production of the FABP5 protein in a human RPE cell line was inhibited using RNA interference technology. As a result, the levels of cholesterol and cholesterol ester were decreased by about 40%, whereas FFAs and triglycerides were increased by 18 and 67% after siRNA treatment, respectively. Some species of phospholipids were decreased in siRNA-treated cells. Cellular lipid droplets were evident and apoB secretion was decreased by 76% in these cells. Additionally, we discovered that ARPE-19 cells could synthesize and secrete Apolipoprotein B100 (apoB100), which may serve as a backbone structure for the formation of lipoprotein particles in these cells. Our results indicate that FABP5 mRNA knockdown results in the accumulation of cellular triglycerides, decreased cholesterol levels, and reduced secretion of apoB100 protein and lipoprotein-like particles. These observations indicated that FABP5 plays a critical role in lipid metabolism in RPE cells, suggesting that FABP5 downregulation in the RPE/choroid complex in vivo might contribute to aging and early age-related macular degeneration.
Similar content being viewed by others
Main
To maintain normal retinal function, retinal pigment epithelial (RPE) cells engulf photoreceptor outer segments (ROS)1, 2, 3, 4 enriched in free fatty acids (FFAs).5 Fatty acid-binding proteins (FABPs) are a family of cytosolic 14- to 15-kDa proteins with high binding affinity for various fatty acids in vitro.6 At least 13 distinct FABPs have been identified, each with its own spectrum of binding affinities and tissue distribution.7 FABPs play an important role in the trafficking of FFAs and other lipids in these tissues.8, 9, 10, 11, 12, 13, 14 However, fundamental questions remain regarding their functional role in lipid metabolism. One of the obstacles to pursuing these questions is the fact that the deletion of particular FABP genes has not led to gross phenotypic changes, most likely because of compensatory overexpression of other members of the FABP family, or even of unrelated fatty acid (FA) transport proteins.15
FABP5 is expressed most strongly in epidermal cells, but is also present in many other tissues including mammary gland, brain, liver, kidney, lung, and adipose tissue.15 However, to our knowledge, there is no information available regarding the functional role of FABP5 in RPE cells. We have previously observed the downregulation of FABP5 by transcriptional analysis in the RPE/choroidal complex in a mouse model of aging and early age-related macular degeneration (AMD).16 As part of our ongoing investigation of the factors that affect early AMD, we hypothesized that FABP5 may play a critical role in lipid metabolism and that its downregulation may contribute to lipid accumulation in RPE cells, an early sign of aging in the eye. To test this hypothesis, we have now used siRNA to inhibit the expression of the FABP5 gene in an established human RPE cell line, ARPE-19, and to assess the effect of its knockdown on lipid metabolism in these cells.
In this study, we found that knockdown of FABP5 mRNA expression resulted in (1) decreased FA uptake, which may result from the decreased production of FABP5 and/or the slowdown in FFA metabolism, (2) decreased cellular cholesterol and cholesterol ester, (3) increased cellular FFAs and triglycerides, and (4) decreased secretion of apoB100 in ARPE-19 cells. These observations indicate that FABP5 plays a critical role in lipid metabolism in ARPE-19 cells, suggesting that FABP5 downregulation in the RPE/choroid complex in vivo16 might contribute to age-related changes and early AMD.
MATERIALS AND METHODS
Cell Culture and FABP5 siRNA Transfection
The routine maintenance of the established, immortalized human RPE cell line, ARPE-19, was carried out as described.17 ARPE-19 cells were grown in six-well culture plates or T75 cm2 culture flasks and maintained in Dulbecco's modified Eagle's medium/F12 (DMEM/F12; Invitrogen Corporation, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen).
A smart pool siRNA or four individual siRNAs from the smart pool (Dharmacon Inc., Lafayette, CO, USA) directed against the human FABP5 mRNA (GenBank Accession No. NM 001444) were used. Nontargeting siRNA#2 siCONTROL (NTC; Dharmacon) was used as a control. Transfection experiments were performed according to the manufacturer's instructions. siRNA in OptiMEM (Invitrogen) was mixed with Lipofectamine 2000 at room temperature for 15 min. Cells plated in 6-well or 96-well plates were treated with siRNA for 6 h, and the toxicity of the siRNA treatment was assessed by Hoechst dye staining (Invitrogen) according to the manufacturer's protocol.
Real-Time RT-PCR and Western Blot Analysis
Total RNA was extracted from cultured ARPE-19 cells using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. The purity of RNA was estimated by the absorbance ratio at 260/280 nm. FABP5 expression was determined by real-time RT-PCR (LightCycler; Roche Diagnostics, Nutley, NJ, USA) as described.18 The following two primer pairs were used for FABP5: 5′-CCTGTCCAAAGTGATGATGG-3′ (sense) and 5′-CAGCATCAGGAGTGGGATG-3 (antisense) and FABP4: 5′-AACCTTAGATGGGGGTGTCC-3′ (sense) and 5′-TGGACCTGACTTCAAGCGTA-3′ (antisense). To measure the potential nonspecific interferon response to siRNA treatment, we assessed the expression of 2′5′-oligoadenylate synthetase 1 and 2 (OAS1 and OAS2), interferon-inducible gene myxovirus resistance 1 (Mx1), interferon-stimulated gene factor 3 (ISGF3g), and interferon-inducible transmembrane proteins (IFITM) by real-time PCR using an Interferon Response Detection Kit (System Biosciences, Mountain View, CA, USA).
PCR products were quantified using the second derivative maximum values and calculated with LightCycler analysis software. Controls without template were included in each run. Expression levels were normalized to the expression level of GAPDH using the primer pair 5′-CGACCACTTTGTCAAGCTCA-3′ (sense) and 5′-AGGGGTCTACATGGCAACTG-3′ (antisense). All PCR products were checked by melting point analysis, and reaction specificity was verified by melting curve analysis. Results are reported as mean±s.d. The Student's unpaired two-tailed t-test was used for statistical analysis, and P values ≤0.05 were considered to be significant.
To investigate the effect of FABP5 mRNA knockdown on FABP5 protein levels, we performed Western blot analysis as described.19 Cells were solubilized with RIPA solution (50 mM Tris-HCl, pH 7.4, with 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, and 1 mM EDTA) containing a protease inhibitor cocktail (Complete Mini; Roche). Cell lysates were centrifuged at 15 000 g for 15 min at 4°C, and the protein concentration of the supernatants was determined using the Quick Start Bradford Dye reagent (Bio-Rad Laboratories, Hercules, CA, USA). Cell lysates (10 μg protein per lane) were separated on a 4–20% SDS–PAGE and electrophoretically transferred to a PVDF membrane (Bio-Red). Membranes were incubated with anti-human FABP5 or FABP4 polyclonal antibody,20 then with secondary anti-rabbit antibody conjugated with horseradish peroxidase (GE-Healthcare, Piscataway, NJ, USA). The signal was detected with an ECL chemiluminescence detection system (GE-Healthcare). Films were scanned with a Molecular Image FX scanner (Bio-Rad Laboratories).
Fatty Acid Uptake Assays
To determine the effect of FABP5 mRNA knockdown on FA uptake in siRNA-treated cells, a QBT FA uptake assay (Molecular Devices, Sunnyvale, CA, USA) was performed according to the manufacturer's protocol. ARPE-19 cells were seeded into 96-well black-wall/clear-bottom plates (BD Biosciences, San Diego, CA, USA). When 60% confluent, the cells were treated with 10 nM siRNA for 6 h, then incubated in DMEM/F12 medium with 1% BSA (FA-free) for 24, 48, and 72 h. The cells were washed twice with Hank's balanced salt solution (HBSS) with 0.2% BSA, and the reaction was quenched by adding a Q-Red.1/4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-S-indacene-3-dodecanoic acid (BODIPY-FA) solution. BODIPY-FA, a palmitic acid derivative, is taken up by cells, but the Q-Red.1 quench reagent is excluded from cells. Therefore, the BODIPY-FA that enters the cells is in an unquenched state, and its fluorescence can be detected. For inhibition and competition experiments, palmitic acid or oleic acid at 0.15–150 μM was directly added to the quencher/BODIPY-FA mixture. Kinetic readings were immediately started, using a Synergy HT microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA) at 25°C. The mean V value (the value of the mean slope) was calculated using KC4 software (Bio-Tek).
Nile Red Staining
Neutral lipid deposits in siRNA-treated cells were detected by Nile Red assays performed as described.21 Stock solutions of Nile Red (Sigma Chemical, St Louis, MO, USA; 0.5 mg/ml in acetone) were prepared and stored at −20°C. The cells were seeded at 3.0 × 103 cells/cm2 in a 96-well black-wall/clear-bottom plate. For oleic acid challenge, the cells were incubated in DMEM/F12 medium supplemented with 0.2 and 0.4 mM oleic acid-BSA (Sigma) for 24 h after they were incubated in DMEM/F12 medium containing 1% BSA for 24 h after siRNA treatment. The cells were then incubated with 100 μl of 1 μM Nile Red with 1% Pluronic F-127 (Sigma) for 4 h at room temperature in the dark, then washed and incubated with HBSS for 16 h. Fluorescence was detected using the Synergy HT microplate reader with the KC4 software. Neutral lipid deposits in these cells were visualized by Zeiss Axiovert 200 microscopy (Carl Zeiss Inc., Thornwood, NY, USA).
Measurement of FFAs, Cholesterol, Cholesterol Esters, and Triglycerides
The levels of FFAs, cholesterol, cholesterol esters, and triglycerides were determined with a FFA quantification kit (BioVision, Mountain View, CA, USA), a cholesterol/cholesteryl ester quantification kit (Biovision), and a triglyceride colorimetric assay kit (Zen-Bio Inc., Research Triangle Park, NC, USA), respectively, according to the manufacturers' protocols. Cells were seeded at 1 × 105/cm2 into six-well plates. After siRNA treatment, the cells were cultured in DMEM/F12 containing 1% BSA for 48 h. The cells were washed with phosphate-buffered saline (PBS), then lysed and processed according to the manufacturers' protocols. The measurements were made with the Synergy HT microplate reader.
Electrospray Tandem Mass Spectrometry
Total lipids from samples were prepared according to a modified Bligh and Dyer procedure.22 Each sample was homogenized at 25°C in 10 volumes of deionized water, then in 3 volumes of 100% methanol-containing 30 mM ammonium formate and vortexed. Four volumes of chloroform were added, and the mixture was vortexed and centrifuged at 1000 g for 10 min. The chloroform layer was removed and analyzed by direct injection into a Sciex 3000 tandem mass spectrometer (Thornhill, ON, Canada). Shotgun lipidomics analysis was done using electrospray ionization tandem mass spectrometry (ESI/MS/MS) using methods similar to those described.22 Samples were injected for 3 min, allowing for the accumulation of mass counts, and the sum of the total counts under each peak was used to quantify each species relative to a standard curve. Phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI) were purchased from Sigma. All solvents and chemicals were analytical grade.
Density Gradient Ultracentrifugation and Negative-Stain Electron Microscopy
Lipoprotein-like particles were prepared from the culture medium as described.23 Cells in T-75 cm2 flasks were treated with 10 nM of siRNA for 6 h, washed with PBS, and incubated with DMEM/F12 medium containing 1% BSA for 48 h. The medium was harvested, and cellular debris was pelleted by centrifugation at 5000 r.p.m. for 10 min at 4°C. The density was adjusted to 1.24 g/ml by adding solid KBr (Sigma). To a centrifuge tube, 10 ml samples were added, overlaid with 2 ml of 1.21 g/ml KBr, and centrifuged at 40 000 r.p.m. for 36 h at 10°C in a Beckman L90 ultracentrifuge using an SW41Ti rotor. Supernatants (0.2 ml) were drawn from the tube top.
Lipoprotein-like particles in the d≤1.21 g/ml fraction were visualized as described.24 In brief, the supernatants were dialyzed into a buffer composed of 0.125 M ammonium acetate, 2.6 mM ammonium carbonate, and 0.26 mM EDTA (pH 7.4) overnight at 4°C, and then mixed with an equal volume of 2% potassium phosphotungstate. For each sample, a 5-μl droplet was placed onto the center of a freshly discharged 400 mesh copper grid (Ted Pella Inc., Redding, CA, USA) with carbon-coated Parlodion support film. After a 5-min absorption, the grids were air dried. The specimens were viewed with a Hitachi H-7600 TEM operating at 80 kV, and images were digitally captured with an AMT 2K camera. The dialysis buffer was examined as a negative control.
35S-Labeled ApoB Immunoprecipitation
ARPE-19 cells were seeded at 1 × 105 cells/cm2 in six-well-plates and grown to 60% confluence, then treated with 10 nM FABP5 siRNA or NTC siRNA for 6 h and incubated in DMEM/F12 medium containing 1% BSA for 24 h. ApoB in the cells and the cultured medium was isolated as described.25 Cells were preincubated for 1 h with methionine- and cysteine-free Dulbecco's modified Eagle's medium (MEM; Sigma), and then for additional 16 h in MEM containing 230 μCi [35S] methionine/cysteine (Amersham Biosciences, Piscataway, NJ, USA). The culture medium was collected and centrifuged. An anti-apoB100 polyclonal antibody (Abcam, Cambridge, MA, USA) was added to the medium and incubated for 16 h at 4°C. Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was then added, and after 30 min the mixture was centrifuged at 13 000 g for 2 min, and isolated 35S-apoB100 was separated on a 4–20% SDS–PAGE gel. The dried gel was exposed to a phosphorimager screen for 16 h before visualization. The specificity of the anti-apoB100 antibody used for 35S-apoB immunoprecipitation was verified by Western blot analysis using human apoB100 (USBiological, Swampscott, MA, USA) as a positive control.
Statistical Analysis
Statistical analysis was performed using Student's t-test or analysis of variance (ANOVA), and P≤0.05 was considered significant. Results are presented as mean±s.d.
RESULTS
Knockdown of FABP5 mRNA by siRNA
To inhibit the expression of the FABP5 gene, we transfected ARPE-19 cells with the smart pool siRNA at 0.001–100 nM. The inhibition of FABP5 gene expression produced by siRNA was dose dependent from 0.001 to 10 nM at 48 h after siRNA treatment; 10 nM siRNA was the most effective concentration, producing ∼80% inhibition of FABP5 expression relative to NTC controls (data not shown). When we compared the four individual siRNAs (designated I–IV) to the smart pool siRNA, we found that siRNA III had an efficacy similar to that of the smart pool siRNA (data not shown), and its inhibitory effect was maintained for at least 96 h (Figure 1a). In addition, the siRNA treatment had no effect on cell viability (data not shown). To decrease the level of off-target effects that can be induced by siRNA, we used 10 nM siRNA III for the remaining studies.
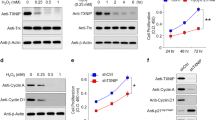
(a) Duration of FABP5 mRNA knockdown by siRNA. ARPE-19 cells were transfected with siRNA at 10 nM FABP5 siRNA, and RNAs were isolated from the samples at 24, 48, 72, or 96 h after siRNA treatment. The expression of the FABP5 gene was determined by real-time RT-PCR. (b) Expression of the FABP4 gene in FABP5 siRNA-treated cells. ARPE-19 cells were transfected with FABP5 siRNA at 10 nM, and RNAs were isolated from the samples at 24 or 48 h after siRNA treatment. All experiments shown in (a) and (b) were repeated at least three times. Bar graphs represent the average of triplicate samples and are presented as mean±s.d. The Student's unpaired t-test was used to compare the differential gene expression between FABP5- and NTC-siRNA-treated cells. * P<0.05.
As FABP4 is very similar in its structure and binding properties to FABP5,20 we determined the mRNA and protein levels of FABP4 at 24 and 48 h after siRNA treatment. FABP4 mRNA and protein levels were not affected by FABP5 mRNA knockdown (Figure 1b; Figure 2b). Western blot analysis, conducted in parallel, shows that FABP5 protein was significantly decreased by 31% at 24 h and by 72% at 48 h after FABP5 siRNA treatment when compared to NTC control (P<0.05; Figure 2a). In contrast, the mRNA (Figure 1b) and protein levels (Figure 2b) of FABP4 at 24 and 48 h after siRNA treatment were not affected.
Effect of FABP5 mRNA knockdown on protein levels of FABP 4 and 5. ARPE-19 cells were treated with 10 nM siRNA and harvested at 24 h and 48 h after siRNA treatment. The protein levels of FABP5 and FABP4 were analyzed by Western blotting. Bands corresponding to FABP4 and 5 were scanned, and the intensity was determined by optical density. The data were normalized by the optical density of GABDH immunostaining in the blots. Representative western blots show the protein levels of FABP5 from NTC- and FABP5-siRNA-treated cells at 24 and 48 h with FABP5 protein as positive control (a), and the protein levels of FABP4 from NTC- and FABP5-siRNA-treated cells at 24 and 48 h with FABP4 protein as positive control (b). The graphs show the mean±s.d, n=3 independent experiments, *P<0.05. Equal loading of samples was normalized by the intensity of GAPDH-immunostaining (c).
siRNA activates a double-stranded RNA activated protein kinase (PKR), causing upregulation of interferon-stimulated genes that may result in nonspecific effects.26 We, therefore, determined whether siRNA treatment induced a nonspecific interferon response with real-time RT-PCR to detect the following markers of a nonspecific immune response: OAS1, ISGF3g,27 and IFITM.28 The expression levels of these genes were unaffected at 24 and 48 h after FABP5 siRNA treatment when compared to NTC controls. Two other markers, Mx1 and OAS2, were not detectable (data not shown), perhaps because of their low expression levels. These results indicate that there was no evidence of siRNA-induced interferon responses in ARPE-19 cells.
Decreased BODIPY-FA Uptake
Liao et al29 reported that BODIPY-FA uptake is closely correlated with the uptake of radiolabeled FAs. To determine the effect of FABP5 mRNA knockdown on the influx of FAs into siRNA-treated cells, we performed BODIPY-FA uptake assays and found that the mean V values for BODIPY-FA uptake at 24, 48, and 72 h after FABP siRNA treatment were decreased by 5%, 8% (P<0.05), and 6% (P<0.05), respectively (Figure 3), indicating that FABP5 mRNA knockdown partially inhibited BODIPY-FA influx into the cells. Furthermore, to examine the specificity of the BODIPY-FA assay, we added palmitic and oleic acids to the assay mixture. As expected, BODIPY-FA uptake was inhibited by both palmitic and oleic acids in a dose-dependent manner, with apparent IC50 (the half maximal (50%) inhibitory concentrations for oleic and palmitic acids of 1.5 and 11.4 μM, respectively (data not shown). These results suggest that BODIPY-FA uptake is mediated by the same mechanism that facilitates the uptake of naturally occurring FAs such as oleic and palmitic acids.
Effect of FABP5 mRNA knockdown on BODIPY-FA uptake. Comparison of kinetic readings of BODIPY-FA uptake by cells was performed at 24 (a), 48 (b), and 72 h (c) after FABP5 siRNA treatment (♦) or NTC siRNA treatment (▪). Changes in fluorescence intensity/area were then plotted over the 60-min time course. The results are given in mini fluorescence units (mFu)/min. Measurements were done in 24-replicate and a Student's t-test was performed.
Increased Cellular Lipid Deposits
FABPs transport FAs to the endoplasmic reticulum for triglyceride and cholesterol synthesis and for lipoprotein assembly,30 we further investigated the effect of FABP5 knockdown on the lipid composition of ARPE-19 cells by measuring cellular cholesterol, cholesterol esters, FFAs, and triglycerides with enzymatic fluorimetric assays after FABP5 mRNA knockdown. We next investigated the effect of FABP5 knockdown on neutral lipid accumulation in siRNA-treated cells by performing the Nile Red binding assay. Although no significant difference in the intensity of Nile Red signal was found between FABP5 siRNA-treated cells and NTC siRNA-treated cells, a slight and reproducible increase in the intensity of the Nile Red staining was seen in FABP5 siRNA-treated cells compared to NTC siRNA-treated controls (Figure 4a). The cells treated with FABP5 siRNA developed large Nile Red-stained lipid deposits compared to NTC control (data not shown).
The levels of neutral lipids measured by Nile Red staining. (a) After treated with siRNA for 6 h, the cells were incubated in the medium containing 1% BSA for 24 h. Nile Red assay was performed after cells incubated in medium supplemented with 0–0.4 mM oleic acid for an additional 24 h. (b) Cytotoxicity assays. The cells were incubated with 10 μM of resazurin in HBSS. The background fluorescence was read twice, with the fluorescence being recorded at 560/590 nm: immediately and after 1 h of incubation at room temperature. Data shown are mean±s.d from four replicates of one representative experiment from three independent experiments. *P<0.05 relative to controls.
To amplify the effect of the knockdown of FABP5 mRNA on neutral lipid deposition, we challenged siRNA-treated cells with oleic acid, which increases the synthesis of triglycerides.30 As a result, the intensity of Nile Red staining was increased in FABP5 siRNA-treated cells (Figure 4a) when incubated in a medium supplemented with 0.2 and 0.4 mM oleic acid, compared to that of NTC siRNA-treated cells. Significant increases of Nile Red staining were found in FABP5 siRNA-treated cells challenged with 0.2 and 0.4 mM oleic acid (P<0.05). No dose dependence from 0 to 0.4 mM oleic acid was found. This could be because OA was thought to reach a maximal effect on cellular lipid accumulation at a concentration of 0.2 mM. In parallel, we also saw a marked increase in the size of lipid deposits in FABP5 siRNA-treated cells compared to NTC control (data not shown). Assessment of cell viability after the Nile Red binding assays showed no significant difference between the cells treated with FABP5 siRNA/oleic acid and NTC siRNA-treated cells (Figure 4b), suggesting that the increase in Nile Red binding in FABP5 siRNA/oleic acid-treated cells was not associated with cell death, and thus reflected a true increase in lipid deposits.31
Changes in Lipid Composition of FABP5 siRNA-Treated Cells
As FABPs transport FAs to the endoplasmic reticulum for triglyceride and cholesterol synthesis and for lipoprotein assembly,30 we further investigated the effects of FABP5 knockdown on the lipid composition of ARPE-19 cells by measuring cellular cholesterol, cholesterol esters, FFAs, and triglycerides with enzymatic fluorimetric assays after FABP5 mRNA knockdown. We found that FFAs and triglycerides were significantly increased by 18 and 67%, respectively, when compared to the values in NTC control (Table 1). In contrast, cellular cholesterol and cholesterol esters decreased significantly by 41 and 38%, respectively (Table 1).
As FAs are a component of individual phospholipids, we next asked whether FABP5 mRNA knockdown alters the composition of individual phospholipids due to the disruption in trafficking of FFA in FABP5 siRNA-treated cells. Cellular phospholipids were analyzed using electrospray tandem mass spectroscopy after FABP5 mRNA knockdown. In general, the levels of phospholipids were decreased in FABP5 siRNA-treated cells compared to NTC control. Among them, the analysis of C16:1 and C16:0 of PC and C16:1 of PS showed significant decreases by 7.5 and 65%, respectively (Table 2).
Lipoprotein-Like Particles Isolated from the Culture Medium of FABP5 siRNA-Treated Cells
To explore whether ARPE-19 cells can produce lipoprotein-like particles and to assess the effect of changes in lipid composition on the size of such particles, we used negative-stain electronic microscopy to visualize particles isolated from the cultured medium. The results of this examination confirmed that ARPE-19 cells could indeed produce lipoprotein-like particles. The particles recovered from the in the d≤1.21 g/ml fraction of the culture medium of siRNA-treated cells were heterogeneous in size and electron density. Although some particles were large and solidly electron lucent, others were small and only slightly electron lucent (data not shown). Particles in the medium in the d≤1.21 g/ml fraction of the medium from NTC siRNA-treated controls were more homogeneous and had an average diameter of 79 nm (range, 44–130 nm), whereas those from FABP5 siRNA-treated cells had an average diameter of 59 nm (range, 33–98 nm). As the variation in the size of the particles was large and the number of particles in the medium was low, we were not able to draw a firm conclusion that the size of the particles was decreased in the medium from FABP5 siRNA-treated cells.
Decreased Secretion of apoB100
The supply of cholesterol available for incorporation into nascent lipoprotein particles may affect apoB secretion by the liver.32, 33, 34 To assess the effect of changes in lipid composition on the secretion of apoB in ARPE-19 cells, we isolated 35S-labeled apoB100 at 520 kDA and its degraded products with apparent MWs of 250 and 75 kDa from the cultured medium by immunoprecipitation. We found that the level of newly synthesized apoB in the culture medium from FABP5 siRNA-treated cells was decreased by 76% (P<0.05) than that from NTC siRNA-treated cells (Figure 5).
Secreted apoB in the medium from siRNA-treated cells. 35S-labeled apoB100 in the medium was isolated by immunoprecipitation, separated on SDS–PAGE, and analyzed by Molecular Image FX. A band with apparent MW of ∼520-kD corresponding to apoB100, and 250-kDa and 75-kDa bands corresponding to degraded apoB100 were detected. The intensity of ∼520-kDa band corresponding to apoB100 was measured. A representative autoradiogram from three independent experiments is shown. *P<0.05.
We used Western blot analysis to validate the apoB secretion data obtained from 35S-labeled apoB100 experiments. We found that secreted apoB in the cultured medium was significantly decreased by 56% in the FABP5 siRNA-treated cells when compared to that of NTC control. This result was consistent with the data obtained from 35S-labeled apoB100 experiments. In addition, the amount of total secreted protein in the cultured medium from FABP5 siRNA-treated cells and that from NTC-siRNA treated cells is comparable under our experimental conditions.
DISCUSSION
In this study, we achieved significant knockdown of FABP5 mRNA and protein (Figures 1 and 2), and verified no changes in the mRNA and protein levels of FABP4, a FABP that is also expressed by ARPE-19 cells, and has functional and structural similarities to FABP5. The specificity of knockdown on the FABP5 mRNA provided an opportunity to identify some of the specific roles of FABP5 in lipid metabolism in ARE-19 cells. This observation is consistent with a report on heterozygous FABP5 knockdown mice, where the expression levels of FABP5 mRNA and protein are decreased by 50% in the brain, skin, and liver without affecting the expression of FABP 1, 3, 4, 6, and 7.35
This study has demonstrated that knockdown of the FABP5 mRNA results in several significant changes in ARPE-19 cells: (1) an inhibition of FA uptake, which may result from the decreased production of FABP5 and/or the slowdown in FFA metabolism, (2) altered cellular lipid composition and an increase in cellular lipid droplets, and (3) decreased secretion of apoB100. As a principal function of FABP5 is to transport FAs in the cytoplasm to organelles, such as the endoplasmic reticulum, the lack of FABP5 protein may disrupt the intracellular transport of FAs, resulting in an 18% increase in cellular levels of FFA in FABP5 siRNA-treated cells compared to that in NTC siRNA-treated cells (Table 1). Such a cellular accumulation of FAs can potentially alter lipolysis and lipogenesis in FABP5 siRNA-treated cells. As most triglycerides are found in lipid droplets within the cytosol and are not associated with the endoplasmic reticulum, triglycerides must be mobilized by lipolysis, followed by reesterification, for triglycerides to be reassembled on the endoplasmic reticulum before being incorporated into lipoprotein particles.36, 37 These processes may have been interrupted by the lack of FABP5, causing a decrease in the efficacy of FA release or blocking the reesterification of liberated FAs in the endoplasmic reticulum. In contrast, overexpression of FABP5 has been reported to increase lipolysis in adipose cells.38
The presence of lipid droplets is an indicator of neutral lipid accumulation, such as FFAs, cholesterol, cholesterol esters, and triglycerides in cells.39 To characterize the nature of the lipid droplets in siRNA-treated cells, we determined cellular levels of FFAs, cholesterol, cholesterol esters, and triglycerides. The significant increase in FFAs and triglycerides and decrease in cholesterol and cholesterol esters, which we observed in FABP5 siRNA-treated cells (Table 1), suggests that cellular lipid droplets in these cells are composed of the increased FFAs and triglycerides. Triglycerides are particularly likely to be involved because they are a major component of cellular neutral lipids, and they were increased by 67% after the knockdown of the FABP5 mRNA. This interpretation is further supported by our observation that the size of the lipid droplets in siRNA-treated cells increased after challenge with oleic acid, which promotes the production of triglycerides.30 The decreased levels of phospholipids in siRNA-treated cells may be caused by the disruption of FAs to be incorporated into phospholipids in the endoplasmic reticulum, resulting from the lack of FABP5. It would be important to investigate the long-term effect of FABP5 mRNA knockdown on RPE lipid metabolism in vivo. Newberry et al reported that liver fatty acid-binding protein (L-FABP) regulates murine hepatic fatty acid trafficking in response to fasting. Their study showed that L-FABP knockout mice fed a high-fat Western diet for up to 18 weeks are less obese and accumulate less hepatic triglyceride than C57BL/6J controls.40
In contrast to the increased levels of FFAs and triglycerides that we observed with FABP5 mRNA knockdown, the levels of cholesterol and cholesterol esters were substantially decreased. One possible explanation for the deceased cellular levels of cholesterol and cholesterol esters is that FAs are important for a variety of cellular processes, including mitochondrial β-oxidation.41 FABPs are thought to transport FAs to the mitochondria for β-oxidation.42 Because of the disruption of intracellular transport caused by FABP5 mRNA knockdown, FAs may not have been transported to the mitochondria for β-oxidation; under these circumstances, the levels of acetyl CoA derived from β-oxidation for cholesterol synthesis would have been expected to decrease, resulting in decreased levels of cholesterol because cholesterol is primarily synthesized from acetyl CoA through the HMG-CoA reductase pathway.43 It has been reported that short-chain FAs (acetic, propionic, and butyric acids) suppress cholesterol synthesis in vivo.44 Therefore, it is possible that some species of the excess FAs that accumulate might inhibit cholesterol synthesis by a mechanism that involves, at least in part, the downregulation of either the activity or expression of the HMG-CoA reductase, a key regulatory enzyme of cholesterol synthesis.
The decreased cholesterol and cholesterol ester composition that we observed as a result of FABP5 mRNA knockdown, led us to examine the secretion of apoB100 in FABP5 siRNA-treated cells. The apoB-containing lipoprotein assembly/secretion pathway is dependent on (1) the production of apoB, which is necessary for the assembly of lipoprotein particles containing a neutral lipid core, (2) the availability of phospholipids (mainly PC) and free cholesterol to form the monolayer surface and triglycerides and cholesterol ester to form the core of the particles; and (3) the availability of microsomal triglyceride transfer protein (MTP).45 In the absence of any one or more of these factors, apoB is diverted from the lipoprotein particle assemble/secretion pathway and is degraded. In HepG2 liver cells, cholesterol ester reduction has been shown to significantly reduce the secretion of apoB-containing lipoproteins.46 Both heparin and lactoferrin were independently able to block the very low density lipoprotein (VLDL)-mediated increases in intracellular cholesteryl ester mass (−56 and −46%) without decreasing triglyceride mass; the also reduced apoB-100 secretion by 53 and 72%, respectively.46 Consistent with these findings, our data suggest that the decreased secretion of apoB100 (Figure 5) could have been caused by a disruption in the formation of apoB100-containing lipoprotein-like particles as a result of a substantial decrease in cholesterol and cholesterol esters after FABP5 knockdown. The lower levels of some species of phospholipids might also contribute the decreased secretion of apoB in siRNA-treated cells (Table 2). In addition, it has been observed that ARPE-19 cells secrete lipoprotein particles in oleic acid-supplemented medium. However, it is not clear that these lipoprotein particles contained apoB.47 In our study, we discovered that ARPE-19 cells could synthesize and secrete apoB100, which may serve as a backbone structure for these lipoprotein particles. We also found that ARPE-19 cells could secrete these lipoprotein particles in physiologic medium. As the variation in the size of the particles was large and the number of particles in the medium was very low, we were not able to draw a firm conclusion that the size of the particles was decreased in the medium from FABP5 siRNA-treated cells.
Our experiments suggest that FABP5 mRNA knockdown may interrupt fatty acid β-oxidation, resulting in decreased cellular cholesterol, cholesterol ester, and some species of phospholipids, the main constituents of lipoprotein-like particles. As a consequence, it may also reduce the production of apoB-containing lipoproteins and lead to decreased secretion of apoB as well as the accumulation of cellular FFAs and triglycerides. One of the obstacles to studies in this area has been the fact that deletion of specific FABP genes has not resulted in gross phenotypic changes, most likely as a result of compensatory overexpression of other members of the FABP family, or even of unrelated FA transport proteins.15 However, using RNA interference technology, we have been able to observe dramatic and unexpected phenotypic changes in FABP5 siRNA-treated ARPE-19 cells. These observations suggest that FABP5 may be a key FA transporter and play a critical role in lipid metabolism in human ARPE-19 retinal pigment epithelial cells. This in turn implies a possible scenario in which the downregulation of FABP5 in the RPE/choroid complex in vivo16 might contribute to aging and early AMD.
Accession codes
References
Gal A, Li Y, Thompson DA, et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet 2000;26:270–271.
Vollrath D, Feng W, Duncan JL, et al. Correction of the retinal dystrophy phenotype of the RCS rat by viral gene transfer of Mertk. Proc Natl Acad Sci USA 2001;98:12584–12589.
Mullen RJ, LaVail MM . Inherited retinal dystrophy: primary defect in pigment epithelium determined with experimental rat chimeras. Science 1976;192:799–801.
Dowling JE, Sidman RL . Inherited retinal dystrophy in the rat. J Cell Biol 1962;14:73–109.
Martin RE, Elliott MH, Brush RS, et al. Detailed characterization of the lipid composition of detergent-resistant membranes from photoreceptor rod outer segment membranes. Invest Ophthalmol Vis Sci 2005;46:1147–1154.
Storch J, Thumser AE . The fatty acid transport function of fatty acid-binding proteins. Biochim Biophys Acta 2000;1486:28–44.
Kingma PB, Bok D, Ong DE . Bovine epidermal fatty acid-binding protein: determination of ligand specificity and cellular localization in retina and testis. Biochemistry 1998;37:3250–3257.
Murphy EJ, Barcelo-Coblijn G, Binas B, et al. Heart fatty acid uptake is decreased in heart fatty acid-binding protein gene-ablated mice. J Biol Chem 2004;279:34481–34488.
Coe NR, Simpson MA, Bernlohr DA . Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. J Lipid Res 1999;40:967–972.
Martin GG, Huang H, Atshaves BP, et al. Ablation of the liver fatty acid binding protein gene decreases fatty acyl CoA binding capacity and alters fatty acyl CoA pool distribution in mouse liver. Biochemistry 2003;42:11520–11532.
Martin GG, Danneberg H, Kumar LS, et al. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J Biol Chem 2003;278:21429–21438.
Atshaves BP, McIntosh AM, Lyuksyutova OI, et al. Liver fatty acid-binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem 2004;279:30954–30965.
Vassileva G, Huwyler L, Poirier K, et al. The intestinal fatty acid binding protein is not essential for dietary fat absorption in mice. FASEB J 2000;14:2040–2046.
Zimmerman AW, Veerkamp JH . New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci 2002;59:1096–1116.
Haunerland NH, Spener F . Fatty acid-binding proteins—insights from genetic manipulations. Prog Lipid Res 2004;43:328–349.
Tian J, Ishibashi K, Ishibashi K, et al. Advanced glycation endproduct-induced aging of the retinal pigment epithelium and choroid: a comprehensive transcriptional response. Proc Natl Acad Sci USA 2005;102:11846–11851.
Handa JT, Reiser KM, Matsunaga H, et al. The advanced glycation endproduct pentosidine induces the expression of PDGF-B in human retinal pigment epithelial cells. Exp Eye Res 1998;66:411–419.
Ishibashi K, Tian J, Handa JT . Similarity of mRNA phenotypes of morphologically normal macular and peripheral retinal pigment epithelial cells in older human eyes. Invest Ophthalmol Vis Sci 2004;45:3291–3301.
Wu T, Chiang SK, Chau FY, et al. Light-induced photoreceptor degeneration may involve the NF kappa B/caspase-1 pathway in vivo. Brain Res 2003;967:19–26.
Simpson MA, LiCata VJ, Ribarik CN, et al. Biochemical and biophysical analysis of the intracellular lipid binding proteins of adipocytes. Mol Cell Biochem 1999;192:33–40.
McMillian MK, Grant ER, Zhong Z, et al. Nile Red binding to HepG2 cells: an improved assay for in vitro studies of hepatosteatosis. In Vitr Mol Toxicol 2001;14:177–190.
Cutler RG, Pedersen WA, Camandola S, et al. Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann Neurol 2002;52:448–457.
Li CM, Presley JB, Zhang X, et al. Retina expresses microsomal triglyceride transfer protein: implications for age-related maculopathy. J Lipid Res 2005;46:628–640.
Forte TM, Nordhausen RW . Electron microscopy of negatively stained lipoproteins. Methods Enzymol 1986;128:442–457.
Boren J, Rustaeus S, Olofsson SO . Studies on the assembly of apolipoprotein B-100- and B-48-containing very low density lipoproteins in McA-RH7777 cells. J Biol Chem 1994;269:25879–25888.
Moss EG, Taylor JM . Small-interfering RNAs in the radar of the interferon system. Nat Cell Biol 2003;5:771–772.
Stark GR, Kerr IM, Williams BR, et al. How cells respond to interferons. Annu Rev Biochem 1998;67:227–264.
Johnson MC, Sangrador-Vegas A, Smith TJ, et al. Cloning and characterization of two genes encoding rainbow trout homologues of the IFITM protein family. Vet Immunol Immunopathol 2006;110:357–362.
Liao J, Sportsman R, Harris J, et al. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J Lipid Res 2005;46:597–602.
Zhang YL, Hernandez-Ono A, Ko C, et al. Regulation of hepatic apolipoprotein B-lipoprotein assembly and secretion by the availability of fatty acids. I. Differential response to the delivery of fatty acids via albumin or remnant-like emulsion particles. J Biol Chem 2004;279:19362–19374.
Crisby M, Kallin B, Thyberg J, et al. Cell death in human atherosclerotic plaques involves both oncosis and apoptosis. Atherosclerosis 1997;130:17–27.
Khan BV, Fungwe TV, Wilcox HG, et al. Cholesterol is required for the secretion of the very-low-density lipoprotein: in vivo studies. Biochim Biophys Acta 1990;1044:297–304.
Khan B, Wilcox HG, Heimberg M . Cholesterol is required for secretion of very-low-density lipoprotein by rat liver. Biochem J 1989;258:807–816.
Bisgaier CL, Essenburg AD, Auerbach BJ, et al. Attenuation of plasma low density lipoprotein cholesterol by select 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors in mice devoid of low density lipoprotein receptors. J Lipid Res 1997;38:2502–2515.
Owada Y, Suzuki I, Noda T, et al. Analysis on the phenotype of E-FABP-gene knockout mice. Mol Cell Biochem 2002;239:83–86.
Francone OL, Griffaton G, Kalopissis AD . Effect of a high-fat diet on the incorporation of stored triacylglycerol into hepatic VLDL. Am J Physiol 1992;263:E615–E623.
Gilham D, Alam M, Gao W, et al. Triacylglycerol hydrolase is localized to the endoplasmic reticulum by an unusual retrieval sequence where it participates in VLDL assembly without utilizing VLDL lipids as substrates. Mol Biol Cell 2005;16:984–996.
Hertzel AV, Bennaars-Eiden A, Bernlohr DA . Increased lipolysis in transgenic animals overexpressing the epithelial fatty acid binding protein in adipose cells. J Lipid Res 2002;43:2105–2111.
Kvilekval K, Lin J, Cheng W, et al. Fatty acids as determinants of triglyceride and cholesteryl ester synthesis by isolated hepatocytes: kinetics as a function of various fatty acids. J Lipid Res 1994;35:1786–1794.
Newberry EP, Xie Y, Kennedy SM, et al. Protection against Western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology 2006;44:1191–1205.
Reddy JK, Hashimoto T . Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr 2001;21:193–230.
Veerkamp JH, Peeters RA, Maatman RG . Structural and functional features of different types of cytoplasmic fatty acid-binding proteins. Biochim Biophys Acta 1991;1081:1–24.
Gibbons GF . Regulation of fatty acid and cholesterol synthesis: co-operation or competition? Prog Lipid Res 2003;42:479–497.
Hara H, Haga S, Aoyama Y, et al. Short-chain fatty acids suppress cholesterol synthesis in rat liver and intestine. J Nutr 1999;129:942–948.
Kang S, Davis RA . Cholesterol and hepatic lipoprotein assembly and secretion. Biochim Biophys Acta 2000;1529:223–230.
Avramoglu RK, Cianflone K, Sniderman AD . Role of the neutral lipid accessible pool in the regulation of secretion of apoB-100 lipoprotein particles by HepG2 cells. J Lipid Res 1995;36:2513–2528.
Li CM, Presley JB, Zhang X, et al. Retina expresses microsomal triglyceride transfer protein: implications for age-related maculopathy. J Lipid Res 2005;46:628–640.
Acknowledgements
This work was supported by NEI EY 14005 (JTH), an AHAF Macular Degeneration Grant (JTH), a Research to Prevent Blindness Clinician Scientist award (JTH), an unrestricted RPB grant, the Intramural Research Program of the National Insitute on Aging, DK053189 (DAB), and gifts from Ric and Sandy Forsythe, The Kwok family the Merlau family, and Aleda Wright.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, T., Tian, J., Cutler, R. et al. Knockdown of FABP5 mRNA decreases cellular cholesterol levels and results in decreased apoB100 secretion and triglyceride accumulation in ARPE-19 cells. Lab Invest 90, 906–914 (2010). https://doi.org/10.1038/labinvest.2009.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2009.33
Keywords
This article is cited by
-
Lycorine induces apoptosis of acute myeloid leukemia cells and inhibits triglyceride production via binding and targeting FABP5
Annals of Hematology (2023)
-
Integrating genome and transcriptome profiling for elucidating the mechanism of muscle growth and lipid deposition in Pekin ducks
Scientific Reports (2017)