Abstract
Pulmonary alveolar microlithiasis (PAM) is a rare autosomal recessive disease caused by mutations in SLC34A2 and characterized by intra-alveolar accumulation of microliths. We diagnosed a case of PAM in a 27-year-old Japanese female and identified a novel mutation in SLC34A2 (c.1390 G>C [G464R] in exon 12).
Similar content being viewed by others
Pulmonary alveolar microlithiasis (PAM) is a rare autosomal recessive disease that is characterized by intra-alveolar accumulation of microliths in the absence of a known calcium metabolism disorder.1 Solute carrier family 34 member 2 (SLC34A2) is a type IIb sodium phosphate co-transporter (NaPi-IIb) that is primarily expressed in alveolar type II cells and is responsible for the uptake of phosphate released from phospholipids during pulmonary surfactant recycling. SLC34A2 is the only known sodium-dependent phosphate transporter expressed in the lung and has a key role in clearing phospholipids from the alveolar space by transporting phosphate ions into alveolar type II cells. Mutations in SLC34A2 that cause impaired function or deficiency could therefore result in local accumulation of phosphate in the alveolar space. In Corut et al.2 first reported that SLC34A2 mutations cause PAM, and subsequent studies have also described mutations in this gene in patients with PAM.3,4 Here, we report a case of PAM with a compound heterozygous mutation in SLC34A2 that includes a novel mutation in exon 12.
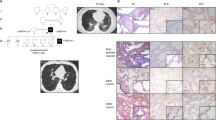
A 27-year-old Japanese female from a non-inbred family presented to our hospital after indication of an abnormal shadow on a chest X-ray was observed during a medical examination. She had no previous medical history and did not present any complaints, such as dyspnea on exertion or cough. A chest X-ray and computed tomography scan showed a diffuse infiltration of fine sand stones in both lungs, mainly in the lower fields and lobes (Figure 1a,b), and the patient was admitted to our hospital for diagnosis of the lung shadow. Physical examinations, including chest auscultation, were normal. Laboratory tests were almost normal and did not suggest the presence of any metabolic disease, sarcoidosis or amyloidosis. Histopathological examination of the lung was performed in the hospital on day 4 and showed lamellar microliths deposited in the alveolar spaces (Figure 1c). On the basis of thoracic imaging and the pathological findings, she was diagnosed with PAM. Furthermore, molecular diagnosis was performed using genomic DNA extracted from the peripheral blood of the patient after obtaining informed consent. Molecular testing on family members was not performed because informed consent could not be obtained. After a 1-year follow-up, the patient had no symptoms and there was no change in the lung shadow.
(a) Chest X-ray and (b) chest computed tomography scan on admission showed a diffused infiltration of fine sand-stones in both lungs mainly in the lower fields and the lower lobe. (c) Hematoxylin and eosin stain of the lung specimen showed lamellar microliths deposited in alveolar spaces (upper; low-power field, below; high-power field).
Genomic DNA was extracted from the peripheral blood of the patient. Using Primer3 software online (Genetyx software, Genetyx, Shibuya, Japan), 12 pairs of primers were designed to amplify SLC34A2 from coding exons 2–13, as well as the intronic flanking sequences. Polymerase chain reaction using a standard protocol was used to amplify the DNA. Direct sequencing was performed for sequencing of polymerase chain reaction products using a 3,500×1 Genetic Analyzer (Thermo Fisher Scientific, Japan). The obtained sequence variants were tested for pathogenicity using Polyphen25 and MutationTaster2.6
As a result of this genetic analysis, a novel compound heterozygous mutation in SCL34A2 was identified in the patient (Figure 2a–d). One mutation, c.1048+1 G>A in intron 9, has already been reported and is causative for PAM.7 A second mutation, c.1390 G>C (G464R), was identified in exon 12. To the best of our knowledge, the latter has not been previously reported. Polyphen2 scores for the novel mutation were 1.0 (sensitivity 0.00, specificity 1.00) using the HumDiv model and 1.0 (sensitivity 0.00, specificity 1.00) using the HumVar model.5 MutationTaster analysis indicated that the novel mutation was a disease-causing mutation (model:simple_aae, prob: 0.999999999912954).6
The patient was initially diagnosed with PAM according to radiological and pathological findings. Mutations in SLC34A2 have been detected in almost all studied patients with PAM; therefore, SLC34A2 is now considered to be the only gene responsible for PAM. We sequenced SLC34A2 to confirm the diagnosis of PAM. SLC34A2 consists of 13 exons, the first of which is non-coding, with the remaining exons encoding a 690-amino acid protein, a type 2 phosphate transporter that is expressed in several human tissues of epithelial origin. As previously detected SLC34A2 mutations were not found within an obvious hotspot region, we sequenced the coding exons and the intronic flanking sequences of SLC34A2. Mutations were detected in intron 9 (c.1048+1 G>A) and exon 12 (c.1390 G>C[G464R]), which strongly supported the diagnosis of PAM. Huqun et al.7 reported that the point mutation c.1048+1 G>A results in aberrant splicing and premature termination of the transcript. Although mutations in exon 12 have been previously reported e.g., the c.1393T>C point mutation8 as well as deletions,9 this case of PAM is the first to be caused by the point mutation c.1390 G>C(G464R). It is unclear how the mutation affects the function of the NaPi-IIb protein, but Polyphen2 and MutationTaster analyses predicted that the novel mutation is detrimental to the protein function.
It was preferable to show that each mutation was located on a different copy of the SLC34A2 gene with either genomic DNA or complementary DNA. However, exons 9 and 12 of the SLC34A2 gene are located on different parts of the genomic DNA; thus, it is very difficult to obtain genomic clones containing both exons 9 and 12 from the patient’s genomic DNA. Furthermore, the mutation in intron 9 was located on the consensus sequence of the splicing donor site and will likely to lead to nonsense decay. Therefore, the isolation of complementary DNA in which intron 9 is incorrectly spliced is considered to be difficult. In all PAM cases in which SLC34A2 mutations were detected, both copies of the SLC34A2 were mutated.2,3,7,9–16 Therefore, it is highly probable that the two mutations observed in this case are considered to affect both alleles.
More than 1,000 cases of PAM have been reported since the first case was described by Malpighi in 1686,4 but to date only 33 patients have been diagnosed with mutations in SLC34A2, with a total of 13 mutant alleles.2,3,7,9–16 Molecular analyses of patients with PAM diagnosed by clinical criteria seem to be important to understand the etiology of PAM and to develop treatments. In conclusion, we identified a case of PAM clinically and genetically and identified a novel heterozygous mutant allele of SLC34A2 that is causative for PAM.
References
References
Tachibana T, Hagiwara K, Johkoh T . Pulmonary alveolar microlithiasis: review and management. Curr Opin Pulm Med 2009; 15: 486–490.
Corut A, Senyigit A, Ugur SA, Altin S, Ozcelik U, Calisir H et al. Mutations in SLC34A2 cause pulmonary alveolar microlithiasis and are possibly associated with testicular microlithiasis. Am J Hum Genet 2006; 79: 650–656.
Vismara MFM, Colao E, Fabiani F, Bombardiere F, Tamburrini O, Alessio C et al. The sodium-phosphate co-transporter SLC34A2, and pulmonary alveolar microlithiasis: presentation of an inbred family and a novel truncating mutation in exon 3. Respir Med Case Rep 2015; 16: 77–80.
Castellana G, Castellana G, Gentile M, Castellana R, Resta O . Pulmonary alveolar microlithiasis: review of the 1022 cases reported worldwide. Eur Respir Rev 2015; 24: 607–620.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Schwarz JM, Cooper DN, Schuelke M, Seelow D . MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–362.
Huqun S, Izumi S, Miyazawa H, Ishii K, Uchiyama B, Ishida T et al. Mutations in the SLC34A2 gene are associated with pulmonary alveolar microlithiasis. Am J Respir Crit Care Med 2007; 175: 263–268.
Proesmans M, Boon M, Verbeken E, Ozcelik U, Kiper N, Van De Casseye W et al. Pulmonary alveolar microlithiasis: a case report and review of the literature. Eur J Pediatr 2012; 171: 1069–1072.
Jönsson ÅLM, Hilberg O, Bendstrup EM, Mogensen S, Simonsen U . SLC34A2 gene mutation may explain comorbidity of pulmonary alveolar microlithiasis and aortic valve sclerosis. Am J Respir Crit Care Med 2012; 185: 464.
Ishihara Y, Hagiwara K, Zen K, Huqun, Hosokawa Y, Natsuhara A . A case of pulmonary alveolar microlithiasis with an intragenetic deletion in SLC34A2 detected by a genome-wide SNP study. Thorax 2009; 64: 365–367.
Dogan O, Ozsahin SL, Gul E, Arslan S, Koksal B, Berk S et al. A frame-shift mutation in the SLC34A2 gene in three patients with pulmonary alveolar microlithiasis in an inbred family. Intern Med 2010; 49: 45–49.
Zhong YQ, Hu CP, Cai XD, Nie HP . A novel mutation of the SLC34A2 gene in a Chinese pedigree with pulmonary alveolar microlithiasis. Chinese J Med Genet 2009; 26: 365–368.
Ma T, Ren J, Yin J, Ma Z . A pedigree with pulmonary alveolar microlithiasis: a clinical case report and literature review. Cell Biochem Biophys 2014; 70: 565–572.
Ozbudak IH, Bassorgun CI, Ozbilim G, Luleci G, Sarper A, Erdogan A et al. Pulmonary alveolar microlithiasis with homozygous c.316g>C (p.G106R) mutation: a case report. Turk Patoloji Derg 2012; 28: 282–285.
Proesmans M, Boon M, Verbeken E, Ozcelik U, Kiper N, Van DCW et al. Pulmonary alveolar microlithiasis: a case report and review of the literature. Eur J Pediatr 2012; 171: 1069–1072.
Wang H, Yin X, Wu D, Jiang X . SLC34A2 gene compound heterozygous mutation identification in a patient with pulmonary alveolar microlithiasis and computational 3D protein structure prediction. Meta Gene 2014; 2: 557–564.
Data Citations
Izumi, Hiroki HGV Database (2016) http://dx.doi.org/10.6084/m9.figshare.hgv.929
Acknowledgements
We acknowledge the patient for her participation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Izumi, H., Kurai, J., Kodani, M. et al. A novel SLC34A2 mutation in a patient with pulmonary alveolar microlithiasis. Hum Genome Var 4, 16047 (2017). https://doi.org/10.1038/hgv.2016.47
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/hgv.2016.47
This article is cited by
-
New insights in the genetic variant spectrum of SLC34A2 in pulmonary alveolar microlithiasis; a systematic review
Orphanet Journal of Rare Diseases (2023)
-
A single-cell atlas of West African lungfish respiratory system reveals evolutionary adaptations to terrestrialization
Nature Communications (2023)
-
Clinical aspects of the phosphate transporters NaPi-IIa and NaPi-IIb: mutations and disease associations
Pflügers Archiv - European Journal of Physiology (2019)