Abstract
Knowledge of the degree of temporal stability of population genetic structure and composition is important for understanding microevolutionary processes and addressing issues of human impact of natural populations. We know little about how representative single samples in time are to reflect population genetic constitution, and we explore the temporal genetic variability patterns over a 30-year period of annual sampling of a lake-resident brown trout (Salmo trutta) population, covering 37 consecutive cohorts and five generations. Levels of variation remain largely stable over this period, with no indication of substructuring within the lake. We detect genetic drift, however, and the genetically effective population size (Ne) was assessed from allele-frequency shifts between consecutive cohorts using an unbiased estimator that accounts for the effect of overlapping generation. The overall mean Ne is estimated as 74. We find indications that Ne varies over time, but there is no obvious temporal trend. We also estimated Ne using a one-sample approach based on linkage disequilibrium (LD) that does not account for the effect of overlapping generations. Combining one-sample estimates for all years gives an Ne estimate of 76. This similarity between estimates may be coincidental or reflecting a general robustness of the LD approach to violations of the discrete generations assumption. In contrast to the observed genetic stability, body size and catch per effort have increased over the study period. Estimates of annual effective number of breeders (Nb) correlated with catch per effort, suggesting that genetic monitoring can be used for detecting fluctuations in abundance.
Similar content being viewed by others
Introduction
Genetic variation is a necessary basis for all levels of biodiversity, but our knowledge of microevolutionary processes and short-term temporal variability patterns of natural populations is limited (Hendry and Kinnison, 1999; Laikre et al., 2005). Studies that monitor genetic variability patterns over time are needed, but most monitoring efforts are restricted to a few years of sampling or single samples separated over relatively long periods of time (Nielsen et al., 1997; Heath et al., 2002; Hansen et al., 2002; Poulsen et al., 2006; Borrell et al., 2008; Hansen et al., 2009; Gomaa et al., 2011). Detailed temporal genetic studies that investigate the genetic composition over microevolutionary time scales—that is, from year to year, cohort to cohort and generation to generation—are accumulating, however (Palm et al., 2003; Dowling et al., 2005; Araki et al., 2007; Palstra et al., 2009; Osborne et al., 2010; Skrbinšek et al., 2012). The representativeness of single samples with respect to genetic composition and level of population variation is also poorly understood. Such information is essential both for understanding how we can interpret information on spatial structures from single samples in time and for designing protocols for how genetic parameters can be monitored (Schwartz et al., 2007).
One parameter, which is often important to monitor, is the effective population size (Ne; Nunney and Elam, 1994; Frankham, 2005; Charlesworth, 2009) because it reflects the rate of inbreeding and amount of genetic variation expected to be lost due to genetic drift each generation. Multiple studies have successfully estimated Ne over time periods separated by several generations. However, for a genetic monitoring program to be effective, it must be able to detect and respond rapidly to possible genetic changes. In these situations, data must be collected continually and analyzed over a continuous and uninterrupted time period. Current knowledge of how Ne fluctuates over short time periods (for example, a few generations) for natural populations is limited. Such knowledge appears to be missing largely because of the practical problems related to conducting temporal studies over multiple generations and the challenge of estimating Ne over short time periods, where accounting for overlapping generations becomes necessary (Jorde and Ryman, 1995; Waples and Yokota, 2007). Similarly, only a few examples exist were Ne has been incorporated into monitoring programs for endangered species or populations (Hansen et al., 2006; Schwartz et al., 2007; Laikre et al., 2008).
Human exploitation of natural populations, including harvesting and large-scale releases, are expected to potentially result in genetic changes of native gene pools, and effects of such operations are particularly important to monitor (Allendorf et al., 2008; Laikre et al., 2010). Salmonid fishes are subject to large-scale commercial and sport fishing and releases worldwide. They are relatively well studied genetically, but information on temporal genetic dynamics for natural, unexploited populations is currently sparse. This information is of importance to permit separation of anthropogenic genetic effects caused by exploitation and releases from what can be attributed to temporal genetic changes in unaffected populations.
In this study, we present data on a lake-resident population of brown trout (Salmo trutta) in central Sweden that has been monitored annually over a 30-year period—1980–2010. Our main objectives are to (i) assess the temporal stability of genetic composition and variation at selectively neutral markers, (ii) estimate Ne over multiple, consecutive generations when accounting for the effect of overlapping generations, (iii) compare these estimates with those obtained when applying an estimator that ignores the effect of overlapping generations and (iv) test for the existence of temporal fluctuations of Ne.
Materials and methods
Study area and sample collection
Lake Blanktjärnen is a small, remote and oligotrophic lake at an elevation of 741 m in the mountain range of the Province of Jämtland, central Sweden (Figure 1). It represents the uppermost part of the river Indalsälven drainage system flowing into the Baltic sea. The lake is eight hectares and shallow (maximum depth 5 m), and brown trout is the only occurring fish species. Blanktjärnen thus typifies thousands of lakes in the Scandinavian mountain range. Together with several other lakes in this area, Lake Blanktjärnen is included in an ongoing genetic monitoring project of brown trout populations. The lake is located in a nature reserve and has been closed for other fishing activities for several decades.
Beginning in 1980, about 100 individuals have been sampled (lethally) each year between June and August, using gillnets of various mesh sizes. Otoliths for age determination and tissue samples for genetic analysis have been collected from each fish, along with information on length, weight, sex and maturity (gonadal development; Jorde and Ryman, 1996; Laikre et al., 1998; Palm and Ryman, 1999; Palm et al., 2003). In this study, individuals collected during 1980–2010 were included, representing a total of 31 sampling years, 37 consecutive cohorts (1972–2008) and 3225 individual fish.
Genotyping
The annual samples have been genotyped for allozyme markers from the start of the project in 1980. Allozymes were long, the only available markers for large-scale genetic screening, and we use them to provide consistency and permit genetic monitoring over an extended period of time. The markers were screened using conventional horizontal starch gel electrophoresis (Allendorf et al., 1977). The following 14 polymorphic loci with a co-dominant gene expression were used in the present study (locus designations follow Shaklee et al. (1990); previous locus designations used by our group are given in brackets to permit comparisons with earlier publications): sAAT-4 [AAT-6], CK-A1 [CPK-1], DIA-1 [DIA], bGALA-2, bGLUA [BGA], G3PDH-2 [AGP-2], sIDDH-1 [SDH-1], sIDHP-1 [IDH-2], LDH-C1 [LDH-5], aMAN, sMDH-2 [MDH-2], ME [MEL], MPI-2 [PMI] and PEPLT.
Statistical treatment
For analyses that are based on collection years, all the 3225 individuals were included. For analyses based on cohorts, we removed cohorts comprising <20 fish, resulting in 3186 individuals representing 32 consecutive cohorts (1975–2006) and sample sizes of 24–190 fish per cohort/locus combination.
Temporal genetic heterogeneity and deviations from Hardy–Weinberg proportions within and among collection years/cohorts, with associated levels of significance, were appraised using CHIFISH (Ryman, 2006) and GENEPOP v. 4.0 (Raymond and Rousset, 1995; Rousset, 2008). Genetic differentiation between sampling years/cohorts was estimated by single- and multi-locus FST values (Weir and Cockerham, 1984). The program STRUCTURE v 2.3.2. (Pritchard et al., 2000; Falush et al., 2003) was used (in addition to Hardy–Weinberg tests) to explore if there are indications of more than a single population in Lake Blanktjärnen. The most likely number of population groupings (clusters, K) compatible with the observed genotypic distributions was tested by individually based likelihood analyses using a burn in length of 500 000 steps (Markov Chain Monte Carlo, MCMC) and 200 000 replicates. Different combinations of assumptions regarding the occurrence of ‘admixture genomes within individuals’ and/or ‘correlated allele frequencies among populations’) were tested (see Pritchard et al., 2007 for details). The number of clusters (K) was set to 1–5, and all individual runs were replicated five times.
The brown trout is characterized by overlapping generations, and therefore unbiased estimates of effective population size (Ne) can currently only be obtained using the method of Jorde and Ryman (1995, 1996). We used this method measuring temporal allele-frequency shifts between consecutive cohorts and applying the unbiased estimator F′s (Fs corrected for sample size) from the TEMPOFS software (Jorde and Ryman, 2007). We obtained an overall Ne estimate for the entire period through averaging the 31 values of F′s from all pairs of consecutive cohorts in the interval 1975–2006. To illustrate the variation of Ne estimates over time, we computed moving averages of F′s based on five and ten consecutive cohorts, respectively.
We constructed a life table for the present population and assessed age-specific survival and reproductive rates following Jorde and Ryman (1996). From this table, we calculated the correction factor (C) and generation interval (G) necessary for correcting for the age-structure effect on allele-frequency shifts (Jorde and Ryman, 1995). Fish were sampled lethally, and we used sample plan II when estimating Ne (Jorde and Ryman, 2007).
We also assessed Ne using the linkage disequilibrium (LD) approach (Waples, 2006) that assumes discrete (nonoverlapping) generations. Ne for single sampling years was assessed using the program LDNE v. 1.31 (Waples and Do, 2008). These separate estimates were used to estimate a weighted harmonic mean Ne for the entire period (1980–2010) as proposed by Waples and Do (2010). Both parametric and jackknife methods were used to obtain 95% confidence intervals, and estimates were calculated assuming random mating and excluding all alleles with frequencies <0.02 (Waples and Do, 2008). Finally, we used LDNE to estimate the annual effective number of breeders (Nb) by analyzing the separate cohorts 1975–2006 (Massa-Gallucci et al., 2010).
The possible existence of deviations from selective neutrality of allozyme alleles was tested by comparing the ratio of the observed variance of temporal allele-frequency shifts among separate loci, measured as Fs between consecutive cohort pairs, to the variance expected when all loci are affected by genetic drift alone (Lewontin and Krakauer, 1973). The logic behind this approach is that under the null hypothesis of selective neutrality, the quantity  , where df=1 in our case because all loci are di-allelic, calculated across loci for each consecutive cohort pair, is expected to follow a Chi-square distribution with df=1.
, where df=1 in our case because all loci are di-allelic, calculated across loci for each consecutive cohort pair, is expected to follow a Chi-square distribution with df=1.
Results
We found no indications of systematic genetic change with respect to population structure, amount of variation or allele frequencies over the study period (Table 1, Supplementary Tables S1–S3). The results are consistent with the existence of one single panmictic brown trout population in Lake Blanktjärnen; there were no significant deviations from Hardy–Weinberg expectations within any of the 31 sampling years, and deviations were found in only 2 of the 32 cohorts (Table 1). Over the total material, there is a weak but statistically significant heterozygote excess with FIS≈−0.01 within both sampling years and cohorts, but we consider this observation of little or no biological significance in light of the small absolute value and the large size of total material underlying the significance testing. As expected from these observations, the STRUCTURE analysis suggested that the genotypic distribution most likely conforms to one population (Pr(K=1)=1.00). Results were consistent between individual runs and over all models examined (Supplementary Table S4).
Gene diversity (expected heterozygosity) ranged in the interval 0.30–0.33 and 0.29–0.35 over those sampling years and cohorts where all loci were scored (Table 1), with no temporal trends (r=0.17, P=0.43 and r=0.23, P=0.26). All the 14 loci were di-allelic and we observed no loss or addition of alleles over time.
Temporal allele-frequency shifts
We detected genetic drift over the study period expressed as highly significant allele-frequency differences among sampling years as well as among cohorts. The overall allele-frequency difference was higher between cohorts (FST=0.010, P<0.001) than between collection years (FST=0.004, P<0.001; Supplementary Table S1) as expected for organisms with overlapping generations (Jorde and Ryman, 1995).
Selective neutrality
No indications of deviation from selective neutrality of alleles over the study period were detected. All the test statistic values fell in the range 0.17–2.19 (mean=0.88±0.16), none over the critical value of 3.84 for one degree of freedom.
Effective population size
Temporal approach: Overall Ne, corrected for overlapping generations based on the present life table data (Supplementary Table S5), was estimated as N̂e=74 (95% confidence interval 50–141; Table 2). We note that negative F′s values result in negative N̂e values, which are interpreted as infinity (N̂e=∞).
We tested for variation in Ne over the period 1975–2006 using a randomization test for equal means of F′s among all pairs of consecutive cohorts and obtained a nonsignificant result (P=0.40). Thus, there are no indications that Ne differs over time based on the entire material. However, restricting the analysis to only include cohorts where all 14 loci were scored yielded a significant result (randomization test for cohorts 1980–2006; P=0.03), and a Kruskall–Wallis test for the same period yielded a result close to nominal significance (P=0.06). A variance analysis on the same material also provides statistical significance (F25, 328 =1.62, P=0.03), and a Tukey honestly significant difference (HSD) post-hoc test suggests that this significance is primarily due to the cohort pairs 1995–1996 and 1999–2000. Thus, we find support for concluding that Ne has not been constant over the study period. Detecting temporal change in Ne using separate cohort pairs is difficult, however, because of the large variation; the estimates of Ne vary in the range 19–∞ (Table 2).
To reduce the effect of variation among individual estimates, we also computed moving averages of N̂e over five and ten consecutive cohorts, respectively. These averages suggest a relatively stable Ne over the first part of the study period with an increase in later years (Figure 2). Averages based on five consecutive cohorts are strongly influenced by estimates from some single cohort pairs, and the two peaks observed are largely due to the infinity estimates observed between cohort pairs 1986–1987, 1999–2000 and 2001–2002 (Figure 2; Table 2). Regressing mean F′s for separate cohort pairs against time indicates that there is a slight tendency of decreasing F′s over time, and thus an increasing Ne (r=−0.15; Supplementary Figure S1), but this tendency is not significant (P=0.42).
Estimates of effective size (temporal method, corrected for age structuring) for the Lake Blanktjärnen population for all pairs of consecutive cohorts (○) and the corresponding mean Ne obtained from moving averages of F′s over five ( ) and ten (
) and ten ( ) consecutive cohorts (cf. Table 2). Cohort on the X-axis represents the first cohort in a pair used for each Ne estimate. The dotted line represents the total estimate for the entire period (N̂e=74; 95% confidence interval 50–141). Note the broken Y-axis and that large Ne estimates are given as numbers (∞=infinity).
) consecutive cohorts (cf. Table 2). Cohort on the X-axis represents the first cohort in a pair used for each Ne estimate. The dotted line represents the total estimate for the entire period (N̂e=74; 95% confidence interval 50–141). Note the broken Y-axis and that large Ne estimates are given as numbers (∞=infinity).
LD approach: Ne estimates for each collection year (1980–2010) with associated 95% confidence intervals are presented in Table 3, and estimates for single sampling years varied in the range 11–∞. Both the weighted and the unweighted harmonic mean Ne estimates are N̂e=76. The weighted estimate assumes than Ne has been constant over the study period, whereas the unweighted harmonic mean does not (Waples and Do, 2010).
Testing for homogeneity of Ne estimates from the LD method is not straightforward, and this issue has not been dealt with explicitly in the literature. We observe no apparent trend over time for the change of the LD estimates, and correlation coefficients are nonsignificant. The general pattern for change of the LD estimates coincides with that of the temporal method, however. The estimates are reasonably stable over the first parts of the study period, increase during the latter part of the period and then return to the initial values (cf. Figures 2 and 3; Tables 2 and 3).
Ne estimates (LD approach, not accounting for age structuring) for single collection years (1980–2010) for the Lake Blanktjärnen population. The dotted line represents the total estimate for the entire period (N̂e=76). Note the broken Y-axis and that large Ne point estimates are given as numbers (∞=infinity).
The estimates of the effective annual number of breeders (Nb; estimated for year t from LD in the cohort born in year t+1) varied in the range 4–∞ (Supplementary Table S6). The overall mean Nb was estimated as N̂b=39 and N̂b=45 for weighted and unweighted harmonic mean, respectively. An increase of N̂b was observed over the period 1983–2006 (r=0.41, P=0.047; Supplementary Figure S2; Supplementary Table S6) when excluding eight early cohorts with low weights and infinity estimates.
Demographic characteristics
The generation length was estimated as G=6.7 from life table data (Supplementary Table S5). No temporal trends in sex ratio were found, and the proportion of males observed for single collection years varied in the range 0.40–0.63 (Table 4). The sex ratio seems to be fairly even over the years, but with a statistically significant excess of males (53%) in the total material (χ21=12.0, P<0.001). The proportion of breeders in single collection years varied in the range 0.08–0.48 and 0.02–0.59 for males and females, respectively, with no significant trend.
Possible changes in length and weight over the study period were examined. We regressed body weight and length against time for 4-year-old fish that represent the largest age group over all years (n>1000). A small but significant increase in both weight and length was observed over the period (weight: r=0.28, P<0.001; length: r=0.27, P<0.001; Supplementary Figures S3 and S4). A significant positive trend was also observed for other age groups (3, 5 and 6-year-old).
There are no indications that the changes in body size are associated with fluctuations of the correction factor C or of the generation interval G used for correcting the Ne estimates for the effect of overlapping generations (C=11.5 and G=6.7; Supplementary Table S5). We constructed life tables for three different time periods (collection years 1980–1989, 1990–1999 and 2000–2010) and calculated C and G for these periods separately obtaining C=15.8, 11.7 and 13.6 and G= 5.9, 6.9 and 6.4 for these periods, respectively. Thus, there is no temporal trend for either quantity. The actual correction factor is C/G (Jorde and Ryman, 1995) that took the values 2.66, 1.70 and 2.12, all being fairly similar without an obvious trend and all resulting in Ne estimates well within the 95% confidence limits of the global estimate.
The number of gillnets used for fish sampling has been recorded from 1988, and we observed a significant increase in the mean number of fish caught per gillnet and night (catch per effort) during 1988–2010 (r=0.77, P<0.001; Supplementary Figure S5). Can this apparent increase of census size be coupled to an increase of the effective number of breeders (Nb)? We found such a positive correlation, and the strongest association is observed between N̂b in year t (estimated from cohort t+1) and catch per effort in t+6 (r=0.63, P≈0.001 based on n=23 x–y pairs) when the progeny from t is 5-year-old and dominates the catch. The correlation between Nb and catch per effort does not appear to be spurious, although both of them are correlated with time; the partial correlation between Nb and catch per effort when keeping time constant is significant (rpartial=0.53, P≈0.01).
Discussion
We conclude that Lake Blanktjärnen, a shallow, oligotrophic lake of eight hectares, harbors a single population of brown trout behaving as one panmictic unit. The genetic composition of this population has remained stable over a 30-year period representing five generations. We observe genetic drift that translates into an overall effective population size of around N̂e=75. The effective size does not appear to have stayed constant over time. There is no obvious temporal trend, but there are indications of an increase of Ne over the latter part of the study period, followed by a decline (Figures 2 and 3). There are indications of increasing body size and abundance.
Estimating effective population size
We are confident that our estimate of overall effective population size is reasonably accurate. We were able to apply the Jorde and Ryman temporal method (1995 and 2007) because demographic information was collected that could be used to assess the parameters C and G. This method is currently the only one that accounts for the effects of overlapping generations and provides an unbiased estimate of Ne (Waples and Yokota, 2007). Detailed life table data is frequently not available, but sensitivity analyses suggest that the temporal method is quite robust to uncertainties in demographic parameter estimates (Jorde and Ryman, 1995, 1996; Palm et al., 2003). Therefore, when demographic information is missing, it can sometimes be extrapolated from other populations with known demography (Heggenes et al., 2009). If age-structure effects are not accounted for, reasonably unbiased estimates are only expected in cases where samples are separated by several generations (Jorde and Ryman, 1995; Waples and Yokota, 2007).
The LD approach for estimating Ne under the assumption of discrete generations provides an overall average estimate that is almost identical to that obtained using the temporal method, that is, N̂e is 74 and 76 for the age-structure corrected temporal and uncorrected LD approaches, respectively. It is not possible to tell, however, whether this similarity of estimates is coincidental or if it reflects a general tendency of the LD approach to be robust to violations of the basic assumption of nonoverlapping generations, because this has not been evaluated (Luikart et al., 2010). Waples and Do (2010) speculate that the LD approach roughly should estimate Ne for a single generation if the number of cohorts represented in the sample is approximately equal to the generation length, although this conjecture needs to be evaluated quantatively (Waples, 2010). In our case N̂e=76 obtained by the LD approach represents a combined estimate over all separate years (Table 3), and collections from specific years in most cases included seven age-classes, which corresponds to the estimated generation time of G=6.7 years.
In semelparous species, such as Pacific salmon (Oncorhynchus spp.), where breeders die after reproduction, Ne can be approximated by the product GNb, where Nb is the harmonic mean annual number of breeders (Waples, 2002, 2010; Wang, 2009). In iteroparous species, like the brown trout, the relationship between Nb and G is more complicated because breeders can reproduce in multiple years, and the relationship between Ne and GNb has not been worked out analytically. In our present material, the weighted harmonic mean estimate of the annual number of effective breeders is N̂b=39 (Supplementary Table S6), yielding a GNb product of ∼260. Thus, although the LD approach applied to a sample of mixed ages results in an estimate of Ne that is very similar to that obtained when correcting for overlapping generations using the temporal method, the LD method results in a substantial downward bias (∼45%) when sampling a single cohort and a gross overestimate (>200%) when using the GNb approximation. Palstra et al. (2009) reported similar results for Atlantic salmon (Salmo salar; iteroparous) and inferred that GNb is generally not a good estimator of Ne in iteroparous species.
Temporal change of Ne
Overall, we found support for Ne not being constant over the study period, but we find no statistical support for a trend over time. The graphical illustrations of Ne estimates over time (Figures 2 and 3) suggest that Ne has been relatively stable over the first part of the study period, followed by an increase and a subsequent return to initial values. Both types of estimates (temporal method and LD) exhibit this pattern but with a time lag when the increase becomes manifest. Such a lag is not unexpected, considering that the estimates obtained by the temporal and LD methods refer to variance and inbreeding effective size, respectively, and that effects of changes in effective size are not expressed simultaneously for these two quantities (Crow and Kimura, 1970, P=361). However, the fact that both methods provide similar patterns of temporal change lends additional support to the notion of true changes of Ne.
Abundance (catch per effort) has increased over time and correlates with the annual effective number of breeders (Nb). This observation suggests that monitoring of Nb can be used for detecting fluctuations of census size in brown trout (see Palstra et al., 2009 and Osborne et al., 2010 for similar observations in other freshwater fishes).
Within our ongoing genetic monitoring project, the effective population size of the Lake Blanktjärnen brown trout has been assessed previously. Jorde and Ryman (1996) estimated Ne for this population to N̂e=97, using a different estimator for genetic drift based on collection years 1980–1993. Further, Laikre et al. (1998) estimated Ne for the female part of the population (Nef) based on the same collection years to N̂ef=52. The estimate presented here adds to the general picture that freshwater resident and relatively isolated brown trout populations normally exhibit quite restricted effective sizes.
Temporal stability of genetic variation
No indication of reduction of genetic variation was found over the 30-year period. Observed and expected heterozygosity remain around 0.3, with the same observed and expected values for the first and last year of study (Table 1). The number of alleles per locus was two for all loci over the entire period, and only in a couple of cases for one locus do we not observe both alleles in the annual sample (Supplementary Table S3). Clearly, with an Ne of around 75, we do not expect to see much reduction of heterozygosity over the five generations, which this period represents, not even in the extreme case of no migration and mutation. Under such conditions, the expected decline of heterozygosity is given by Ht=(1–1/2Ne)t Ht=0 (where t is time in generations and H is heterozygosity), and we would only expect a reduction from 0.31 to 0.27, but it is unlikely that we would be able to detect such a minor change with the current set of loci.
Over longer periods of time, we would expect heterozygosity to decline if the population is isolated, however, and the fact that variation is still relatively high indicates that at least some migration occurs in this system. This is in line with the observation of Jorde and Ryman (1996), who noted quite different Ne estimates for brown trout from different lakes in this area (N̂e from 52 to 480) in spite of very similar levels of heterozygosity. Thus, a small amount of gene flow from neighboring populations appears to be an important factor for maintaining genetic variation within separate lakes (cf. Hindar et al., 2004).
Demographic changes and effects of sampling
We find indications that body size and abundance have increased over time. We have not monitored census size (NC), but we do have one estimate from a mark recapture study conducted in 2009 when NC was estimated as 576 (95% confidence interval 493–706) for fish with a total body length of ⩾19 cm (Charlier et al., 2011). This estimate translates into an Ne/NC ratio of 0.13, which is in line with observations from many other species including salmonids (Frankham, 1995; Palstra and Ruzzante, 2008; Palstra et al., 2009). Assuming that our mean catch per effort reflects an increase of NC, the ratio might have decreased over the study period.
Body size is expected to increase as an effect of fishing if harvest results in a reduced abundance. We collected lethally ∼100 fish per year, which in the light of the NC estimate of <600 appears a quite large proportion of the total population, but there are no indications of a declining population size. Further, there is no indication that the sampling has reduced genetic variation. We see no apparent explanation to our observation of increased abundance, but note that the mean annual temperature has increased over the study period by >1 °C (Swedish Metrological and Hydrological Institute, 2011). Increased fish abundance is a possible effect of increased temperature (Elliott, 1994), but additional studies are required to settle if this is the case for Lake Blanktjärnen.
Genetic monitoring
The basic genetic characteristics (that is, expected heterozygosity, number of alleles per locus, genotypic proportions and population structure) of this population are stable both over short periods of time, that is, from year to year, cohort to cohort and generation to generation, and over decades and almost five generations. Single samples thus provide good estimates of such characteristics in this particular case.
The rate of genetic drift fluctuates over time, however, and had we only sampled parts of this period, we could have obtained a nonrepresentative picture of the effective population size of this population. For instance, the amount of drift appears weaker during the later part of the study period, and Ne would have been estimated as 159 if we only had sampled the period 2005–2010. With this estimate alone, we would have misjudged the rate of drift over longer periods of time, and with our single assessment of census size, we would have estimated the Ne/NC ratio as 0.28, that is, considerably larger than the present estimate of 0.13.
DATA ARCHIVING
Data have been deposited at Dryad: doi:10.5061/dryad.189f4.
References
Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N (2008). Genetic effects of harvest on wild animal populations. Trends Ecol Evol 23: 327–337.
Allendorf FW, Mitchell N, Ryman N, Ståhl G (1977). Isoenzyme loci in brown trout (Salmo-trutta-L) - detection and interpretation from population data. Hereditas 86: 179–189.
Araki H, Waples RS, Ardren WR, Cooper B, Blouin MS (2007). Effective population size of steelhead trout: influence of variance in reproductive success, hatchery programs, and genetic compensation between life-history forms. Mol Ecol 16: 953–966.
Borrell YJ, Bernardo D, Blanco G, Vazquez E, Sanchez JA (2008). Spatial and temporal variation of genetic diversity and estimation of effective population sizes in Atlantic salmon (Salmo salar, L.) populations from Asturias (Northern Spain) using microsatellites. Cons Gen 9: 807–819.
Charlesworth B (2009). Effective population size and patterns of molecular evolution and variation. Nat Rev Gen 10: 195–205.
Charlier J, Palmé A, Laikre L, Andersson J, Ryman N (2011). Census (NC) and genetically effective (Ne) population size in a lake-resident population of brown trout Salmo trutta. J Fish Biol 79: 2074–2082.
Crow JF, Kimura M (1970) An Introduction to Population Genetics Theory. Alpha Editions: Minneapolis, MN.
Dowling TE, Marsh PC, Kelsen AT, Tibbets CA (2005). Genetic monitoring of wild and repatriated populations of endangered razorback sucker (Xyrauchen texanus, Catostomidae, Teleostei) in Lake Mohave, Arizona-Nevada. Mol Ecol 14: 123–135.
Elliott JM (1994) Quantitative Ecology and the Brown Trout. Oxford University Press: New York.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Frankham R (1995). Effective population size/adult population size ratios in wildlife: a review. Genet Res 66: 95–107.
Frankham R (2005). Genetics and extinction. Biol Cons 126: 131–140.
Gomaa NH, Montesinos-Navarro A, Alonso-Blanco C, Picó FX (2011). Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Mol Ecol 20: 3540–3554.
Hansen MM, Fraser DJ, Meier K, Mensberg KLD (2009). Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Mol Ecol 18: 2549–2562.
Hansen MM, Nielsen EE, Mensberg KLD (2006). Underwater but not out of sight: genetic monitoring of effective population size in the endangered North Sea houting (Coregonus oxyrhynchus). Can J Fish Aquat Sci 63: 780–787.
Hansen MM, Ruzzante DE, Nielsen EE, Bekkevold D, Mensberg KLD (2002). Long-term effective population sizes, temporal stability of genetic composition and potential for local adaptation in anadromous brown trout (Salmo trutta) populations. Mol Ecol 11: 2523–2535.
Heath DD, Busch C, Kelly J, Atagi DY (2002). Temporal change in genetic structure and effective population size in steelhead trout (Oncorhynchus mykiss). Mol Ecol 11: 197–214.
Heggenes J, Røed KH, Jorde PE, Brabrand Å (2009). Dynamic micro-geographic and temporal genetic diversity in vertebrates: the case of lake-spawning populations of brown trout (Salmo trutta). Mol Ecol 18: 1100–1111.
Hendry AP, Kinnison MT (1999). The pace of modern life: measuring rates of contemporary microevolution. Evolution 53: 1637–1653.
Hindar K, Tufto J, Sættem LM, Balstad T (2004). Conservation of genetic variation in harvested salmon populations. ICES J Mar Sci 61: 1389–1397.
Jorde PE, Ryman N (1995). Temporal allele frequency change and estimation of effective size in populations with overlapping generations. Genetics 139: 1077–1090.
Jorde PE, Ryman N (1996). Demographic genetics of brown trout (Salmo trutta) and estimation of effective population size from temporal change of allele frequencies. Genetics 143: 1369–1381.
Jorde PE, Ryman N (2007). Unbiased estimator for genetic drift and effective population size. Genetics 177: 927–935.
Laikre L, Jorde PE, Ryman N (1998). Temporal change of mitochondrial DNA haplotype frequencies and female effective size in a brown trout (Salmo trutta) population. Evolution 52: 910–915.
Laikre L, Larsson LC, Palmé A, Charlier J, Josefsson M, Ryman N (2008). Potentials for monitoring gene level biodiversity: using Sweden as an example. Biodivers Conserv 17: 893–910.
Laikre L, Palm S, Ryman N (2005). Genetic population structure of fishes: Implications for coastal zone management. Ambio 34: 111–119.
Laikre L, Schwartz MK, Waples RS, Ryman N (2010). Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends Ecol Evol 25: 520–529.
Lewontin RC, Krakauer J (1973). Distribution of gene frequency as a test of theory of selective neutrality of polymorphisms. Genetics 74: 175–195.
Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW (2010). Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Cons Gen 11: 355–373.
Massa-Gallucci A, Coscia I, O′Grady M, Kelly-Quinn M, Mariani S (2010). Patterns of genetic structuring in a brown trout (Salmo trutta L.) metapopulation. Cons Gen 11: 1689–1699.
Nielsen EE, Hansen MM, Loeschcke V (1997). Analysis of microsatellite DNA from old scale samples of Atlantic salmon Salmo salar: a comparison of genetic composition over 60 years. Mol Ecol 6: 487–492.
Nunney L, Elam DR (1994). Estimating the effective population-size of conserved populations. Cons Biol 8: 175–184.
Osborne MJ, Davenport SR, Hoagstrom CW, Turner TF (2010). Genetic effective size, Ne, tracks density in a small freshwater cyprinid, Pecos bluntnose shiner (Notropis simus pecosensis). Mol Ecol 19: 2832–2844.
Palm S, Laikre L, Jorde PE, Ryman N (2003). Effective population size and temporal genetic change in stream resident brown trout (Salmo trutta, L.). Cons Gen 4: 249–264.
Palm S, Ryman N (1999). Genetic basis of phenotypic differences between transplanted stocks of brown trout. Ecol Freshw Fish 8: 169–180.
Palstra FP, O′Connell MF, Ruzzante DE (2009). Age structure, changing demography and effective population size in Atlantic salmon (Salmo salar). Genetics 182: 1233–1249.
Palstra FP, Ruzzante DE (2008). Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol 17: 3428–3447.
Poulsen NA, Nielsen EE, Schierup MH, Loeschcke V, Grønkjær P (2006). Long-term stability and effective population size in North Sea and Baltic Sea cod (Gadus morhua). Mol Ecol 15: 321–331.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Pritchard JK, Wen X, Falush D (2007) Documentation for STRUCTURE software: version 2.2 Software available from http://pritch.bsd.uchicago.edu/software.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Rousset F (2008). GENEPOP ′ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Res 8: 103–106.
Ryman N (2006). CHIFISH: a computer program testing for genetic heterogeneity at multiple loci using chi-square and Fisher’s exact test. Mol Ecol Notes 6: 285–287.
Schwartz MK, Luikart G, Waples RS (2007). Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22: 25–33.
Shaklee JB, Allendorf FW, Morizot DC, Whitt GS (1990). Gene nomenclature for protein-coding loci in fish. Trans Am Fish Soc 119: 2–15.
Skrbinšek T, Jelenčič M, Waits L, Kos I, Jerina K, Trontelj P (2012). Monitoring the effective population size of a brown bear (Ursus arctos) population using new single-sample approaches. Mol Ecol 21: 862–875.
Swedish Metrological and Hydrological Institute (2011). http://www.smhi.se/klimatdata/meteorologi/dataserier-2.1102.
Wang J (2009). A new method for estimating effective population sizes from a single sample of multilocus genotypes. Mol Ecol 18: 2148–2164.
Waples RS (2002). Effective size of fluctuating salmon populations. Genetics 161: 783–791.
Waples RS (2006). A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Cons Gen 7: 167–184.
Waples RS (2010). Spatial-temporal stratifications in natural populations and how they affect understanding and estimation of effective population size. Mol Ecol 10: 785–796.
Waples RS, Do J (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Res 8: 753–756.
Waples RS, Do J (2010). Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3: 244–262.
Waples RS, Yokota M (2007). Temporal estimates of effective population size in species with overlapping generations. Genetics 175: 219–233.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Acknowledgements
We thank Per Erik Jorde and Robin Waples for their helpful discussions and Anna Palmé and two anonymous reviewers for their valuable comments on an earlier version of this paper. The study area is located in the Hotagen Nature Reserve, Province of Jämtland, Sweden, and we acknowledge the cooperation from the Jiingevaerie Sami Community and the Jämtland County Administrative Board. We also greatly appreciate the help from voluntary field workers over the years. LL and NR acknowledge financial support from The Swedish Research Council, The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) and the BONUS Baltic Organizations’ Network for Funding Science EEIG (the BaltGene research project). LL also acknowledges support from the Carl Trygger Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Charlier, J., Laikre, L. & Ryman, N. Genetic monitoring reveals temporal stability over 30 years in a small, lake-resident brown trout population. Heredity 109, 246–253 (2012). https://doi.org/10.1038/hdy.2012.36
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.36
Keywords
This article is cited by
-
Monitoring genome-wide diversity over contemporary time with new indicators applied to Arctic charr populations
Conservation Genetics (2024)
-
Long-term monitoring of a brown trout (Salmo trutta) population reveals kin-associated migration patterns and contributions by resident trout to the anadromous run
BMC Ecology and Evolution (2021)
-
Reproductive dynamics of a native brook trout population following removal of non-native brown trout from a stream in Minnesota, north-central USA
Hydrobiologia (2019)
-
Complex genetic diversity patterns of cryptic, sympatric brown trout (Salmo trutta) populations in tiny mountain lakes
Conservation Genetics (2017)
-
Genetic assessment of Abies koreana (Pinaceae), the endangered Korean fir, and conservation implications
Conservation Genetics (2017)

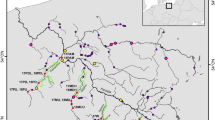

 ) and schematic representation of Lake Blanktjärnen and connected water systems. Small arrows indicate the direction of waterflow and numbers represent elevations (m).
) and schematic representation of Lake Blanktjärnen and connected water systems. Small arrows indicate the direction of waterflow and numbers represent elevations (m).
